How to Cite | Publication History | PlumX Article Matrix
Ujwala Vinayak Khisti1, Suyash Arun Kathade2, Mayur Arjun Aswani2, Pashmin Kaur Anand2 and Nirichan Kunchirman Bipinraj*2
1Department of Microbiology, Annasaheb Magar College, Hadapsar, Pune, India.
2Bharati Vidyapeeth Rajiv Gandhi Institute of I.T. and Biotechnology, Katraj, Pune, India.
Corresponding Author E-mail: bipinrajnk@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2735
ABSTRACT: Probiotics are live microorganisms which upon ingestion confer health benefits to the host and are widely applied for human and animal welfare. The present study reports the isolation of yeast cells from caterpillar frasses and its probiotic characterization. Out of four yeast cultures isolated, all found to be non-hemolytic and cultures designated as CV-I, CV-II CV-III and CV-IV showed good bile tolerance at 1.2%. These cultures possessed the ability to grow pH range of 1.5 – 10, exhibited auto-aggregation and co-aggregation capabilities, which are essential for growth in alimentary canal and reduction of pathogen adherence on the intestinal epithelial cells. All cultures exhibited good tolerance to temperature up to 42°C. Isolate CV-I showed wide range of antimicrobial activities against pathogenic bacteria and fungi. This study is the first report of isolation and characterization of probiotic yeast from caterpillar frass. The isolate CV-I has been identified as Saccharomyces cerevisiae by molecular methods. This culture is an ideal candidate for further probiotic exploration.
KEYWORDS: Azadirachta Indica; Caterpillar Frass; Probiotic Yeast; Saccharomyces Cerevisiae
Download this article as:| Copy the following to cite this article: Khisti U. V, Kathade S. A, Aswani M. A, Anand P. K, Bipinraj N. K. Isolation and Identification of Saccharomyces Cerevisiae from Caterpillar Frass and Their Probiotic Characterization. Biosci Biotech Res Asia 2019;16(1). |
| Copy the following to cite this URL: Khisti U. V, Kathade S. A, Aswani M. A, Anand P. K, Bipinraj N. K. Isolation and Identification of Saccharomyces Cerevisiae from Caterpillar Frass and Their Probiotic Characterization. Biosci Biotech Res Asia 2019;16(1). Available from: https://bit.ly/2I3JrSv |
Introduction
According to FAO/WHO 2001 reports, probiotics are stated as live micro-organisms which when administered in adequate amounts, confer a health benefit on host, such as production of antimicrobial compounds, modulation of immune response, confer resistance to food antigens, assimilate cholesterol, prevent autoimmunity etc.1 They also posses the ability to enhance digestion and utilization of nutrients,2 along with production of precursors of aroma compounds such as free amino acids, free fatty acids etc.3 Considering these benefits, probiotics are increasingly used in commercial animal production, thereby improving animal health and productivity. According to ICMR-DBT, WHO and world gastroenterology organization reports a probiotic strains should not be toxic, it should tolerate wide range of pH and temperature and should be able to tolerate the gastric and intestinal environment. Although many yeasts and bacteria are characterized as a probiotic organism, still there is scope for exploring different sources for probiotic microorganisms. Gut microflora is very critical for the health and survival for any organism. It helps the organism to digest the food, detoxify the alkaloids and improve the immunity.4 Hence, it is likely to isolate a beneficial microorganism from the gut of a healthy organism. This study reports for the first time the isolation of probiotic yeast from the frass of caterpillar of Indarbela quardinotata collected from bark of Azadirachta indica a very common and widely used medicinal plant.
Material and Methods
Culture Isolation
Caterpillars (Indarbela quardinotata) were collected from bark of Azadirachta indica tree in Pune, India. The caterpillars were washed 3 times with sterile distilled water to reduce surface contamination. Washed caterpillars were kept in sterile petriplate and frass (0.5 g) samples were collected from petriplate after 24 h. The samples (0.5 g) were made into a suspension with sterile saline (0.8%). It was then streaked in YPD medium (Yeast extract Peptone Dextrose in 1:2:2 wt/Vol), pH 6.5 (Himedia, India) for isolation of yeast cells. The plates were incubated at 37°C for 48 h. After incubation, morphologically distinct colonies were isolated and observed microscopically. Cells that showed yeast morphology were then purified and stored in agar slants (YPD). These cultures were further inoculated in YPD broth for 24 h., centrifuged and suspended in saline to get 107cfu/ml. This suspension (1%) was used as inoculum in all experiments.
Toxicity Assay
For toxicity assay isolates were spot inoculated on sheep blood agar plates and incubated at 37°C for 24 h. Toxicity was determined by pattern of hemolysis on blood agar plates.5
pH Tolerance
For pH tolerance study, media pH was adjusted to 1.5 to 10 using 1N HCl or 1N NaOH before inoculating the culture. After inoculation the media were incubated at 37°C for 24 h. and observed for growth.6
Bile Tolerance
YPD broth containing different concentrations of bile salts (Loba Chem, India) 0.3, 0.6, 0.9 and 1.2% were inoculated with culture suspension and growth was compared with bile less control after incubation at 37°C for 24 h.6
Temperature Tolerance
Cultures were incubated in YPD broth and incubated at three different temperatures at 28°C, 37°C and 42°C. Growth was observed spectrophotometrically at 600nm (Thermo Scientific) after 24 h. incubation.6
Simulated Gastric and Intestinal Juice Tolerance Assay
To determine in-vitro survival of the isolates, culture suspension in PBS (0.2 ml) were added in mixture of 1 ml gastric juice pepsin (pH 2) or pancreatin (pH 8) and 0.3 ml of sodium chloride (0.5% w/v) as described by.7 The gastric and intestinal transit tolerance was evaluated by viable count of the cultures checked at 1, 90 and 180 min for gastric and 1, and 240 min for intestinal juice by spreading the suspension on YPD agar plates.
Auto-aggregation and Co-aggregation Assay
Cultures were grown in YPD broth for 24 h. at 37°C. Cell pellets were obtained by centrifuging it at 5000 rpm for 5 min and suspended in 2ml of sterile PBS (pH 7.4). Cell suspension (1ml) was added to 4ml of sterile PBS solution in a test tube and was vortexed for 10 seconds and was incubated for 24h. at 37°C. Sample (1ml) was carefully taken at 0, 2, 4 and 24 h. and the optical density was determined at 600 nm. Co-aggregation was estimated as described by.8 Accordingly, culture were suspended in 0.2 ml of PBS and mixed with equal amount of pathogen (0.5 OD) and the final volume were adjusted to 5ml. It was then incubated for 24 h. at 37°C. The OD at 600 nm of the suspension were measured (2, 4 and 24 h.) and compared with pathogen suspension. After 24 h of incubation, yeast cultures were taken and stained by methylene blue and observed under microscope.9
Antimicrobial Assay
Antimicrobial assays was carried against enteropathogens such as; Escherichia coli, Staphylococcus aureus, Enterococcus fecalis, Candida albicans 3557. The pathogens were spread plated on Muller Hinton agar and plates were incubated at 37°C for 10 min. Wells (5mm) were punctured in agar using punch borer. The isolates were grown in YPD broth at 37°C for 24 h. The supernatant was collected after centrifuging at 10000 rpm for 20 min at room temperature and 20µl were added into the agar well. Inhibition zones were measured after incubating it for 24 h. at 37°C.9,10
Molecular Identification
Total genomic DNA was isolated from pure and isolated colonies as the method described by Polkade A.V. et.al., 2015.11 Culture was grown in YPD broth and centrifuged to obtain cell pellets. Cell pellets were suspended in extraction buffer (100 mM Tris/HCl pH 8.0; 100 mM Na2EDTA, pH 8.0), Proteinase K (Invitrogen) was added to final concentration of 100 mg/ml and mixture was incubated at 55°C for 2 hr. in shaking condition. DNA was extracted by phenol:chloroform:isoamyl alcohol (25:24:1). Extracted DNA was washed with 70% ethanol and dissolved in Tris-EDTA buffer (pH 8.0). The ITS region of DNA was amplified by using primers ITS1 TCCGTAGGTGAACCTGCGG and ITS4 TCCTCCGCTTATTGATATGC. Strain was identified by amplification and sequencing of ribosomal Internal Transcribed Spacer (ITS) region. PCR products were purified by PCR Cleanup Kit (Qiagen). Purified amplicons were sequenced on both strands on an ABI 3730xl DNA analyser using the Big Dye terminator kit (Applied Biosystems). The sequences were assembled using DNA star Pro and analysed for their closest relatives based on pairwise sequence similarity by using Blast Logarithmic Alignment Search Tool (BLAST) algorithm in National Centre for Biotechnology Information (NCBI) database (minimum 97% sequence similarity). Each experiment was performed in triplicate and mean value were calculated.
Results and Discussions
Isolation of Yeast Cultures
YPD medium was used for yeast isolation and total 42 morphologically distinct colonies were appeared in the isolation plates. These cultures were further screened by Gram staining and negative staining to observe yeast cells. Out of total cultures only four cultures, designated as CV-I, CV-II, CV-III and CV-IV showed yeast cell morphology and selected for further studies.
Toxicity Assay
Hemolysis was recorded in three forms α, β and γ lysis. While α and β indicate toxicity γ is non-lysis and hence non-toxic. All four cultures tested showed γ lysis on sheep blood agar plate and hence eligible for further probiotic characterization.
Tolerance to pH and Bile Salt
Generally, probiotics are administered through oral route, so it is utmost important to survive against harsh acidic and alkaline conditions present in stomach and intestine respectively(9,10). In the present study, all cultures were able to survive in alkaline condition up to pH 10, while it showed fair growth in pH 2.5 and 3.5 and excellent growth in neutral range of pH 7.
All cultures were able to survive in bile salt concentrations of 0.9% showing good growth. Out of four isolated yeast cultures, CV-I and CV-II showed good growth even in bile concentration of 1.2%. Certain reports suggest that Sacchromyces boulardii did withstand bile concentration of 1.2% even after 48 h. of incubation. After passage through the acidic stomach conditions, it is important that, probiotic strains are able to survive the bile salt in the intestine normal concentration of bile around 0.3% to 2%.12
Effect of Temperature
All yeast isolates were able to grow at three different temperatures 28°C, 37°C and 42°C. It was found that CV-II had good growth overall at three different temperature reading the O.D.at 0.367, 0.456 and 0.577 at 28°C, 37°C and 42°C respectively at 600nm.
Gastric and Intestinal Tolerance
This experiment is aimed to find out the resistance of cultures to gastric and intestinal juices. An ideal culture should survive 2 h. of exposure to gastric and intestinal juice of pH 2. It is detected by TVC after exposure. Present isolates showed varying degrees of resistance when exposed to pepsin and pancreatin. All tested cultures, CV-I, CV-II, CV-III, CV-IV, were found to be surviving in both conditions, however, the cell count decreased after incubation up to 240 min (Figure 1 & 2). Studies by13 showed that known probiotics such as Lactobacillus and Bifidobacterium, after the treatment of such harsh enzymes and at extreme pH conditions down regulated their growth similar to this study.
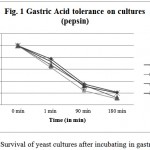 |
Figure 1: Survival of yeast cultures after incubating in gastric juice.
|
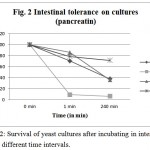 |
Figure 2: Survival of yeast cultures after incubating in intestinal juice at different time intervals.
|
Auto-aggregation and Co-aggregation
Auto-aggregation percentage of yeast isolates was measured by comparing the initial absorbance at 600 nm. Minimum required auto-aggregation percentage for good probiotic isolates has been recommended more than 40%. CV-I, CV-II showed better aggregation than CV-III, CV-IV (Figure 3). From the graph it is clear that CV-I and CV-II showed 100% aggregation after 24 h., were as CV-III and CV-IV showed only 65.55% aggregation after 24 h.
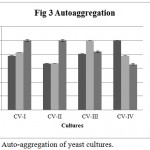 |
Figure 3: Auto-aggregation of yeast cultures.
|
Co-aggregation percentage of yeast isolates were measured by absorbance and microscopic observation. Co-aggregation is the measure of the ability to inhibit the pathogenic microorganism. When exposed to E. coli, CV-IV showed 100% co-aggregation ability after 24 h. (Figure 4) were as CV-I (96%), CV-II (92.68%) and CV-III (70.93%) showed comparatively lesser ability. Similarly, the isolates could co-aggregate with E. fecalis (CV-I, CV-II, CV-III, CV-IV : 100% aggregation, Figure 5) and S. aureus (CV-I, CV-II and CV-IV: 100, Figure 6) after 24 h. indicating they are very good in controlling these pathogens.
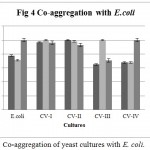 |
Figure 4: Co-aggregation of yeast cultures with E. coli.
|
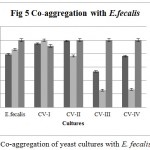 |
Figure 5: Co-aggregation of yeast cultures with E. fecalis.
|
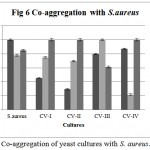 |
Figure 6: Co-aggregation of yeast cultures with S. aureus.
|
For a culture to be probiotic, the first mandatory criterion is non-toxicity or safety of culture for host (WHO-FAO). Generally, probiotics are administered through oral route, so it is utmost important to survive against harsh acidic and alkaline conditions present in stomach and intestine respectively.13,14 After passage through the acidic stomach conditions, it is important that, probiotic strains able to survive the bile salt in the intestine normal concentration of bile around 0.3% to 2%.15 Present study showed the four isolates had better growth at 42°C followed by 37°C and 28°C after 24 hour of incubation. However, in vitro probiotic study by Lohith and Anu (2014) showed there was reduced growth of probiotic yeasts at 42°C indicating present yeasts isolates had ability to tolerate higher temperatures. Study by Charteris et al (1998) showed that known probiotics such as Lactobacillus and Bifidobacterium, after the treatment of such harsh enzymes and at extreme pH conditions down regulated their growth similar to this study. Study by Ogunremi et.al., 2015 showed auto-aggregation of probiotic yeasts cultures increased with increase in time, however in present report CV-I and CV-II cultures showed similar result whereas CV-III and CV-IV cultures indicated decrease in aggregation capabilities. Co-aggregation abilities, showed good aggregation abilities with pathogens E.coli, E.fecalis and S.aureus showed similar result with Ogunremi et.al., 2015. Auto-aggregation is an indication of the probiotic cultures’ ability to adhere on the intestinal epithelium, which leads to colonization. Similarly, co-aggregation indicates the ability to inhibit pathogen from adhering on the epithelium. All four isolates in the present study showed high percentage of auto and co-aggregation.
Antimicrobial Activity
Bioactive compounds present in supernatant were tested against Gram positive and Gram negative bacteria. The results depicted that CV-I, CV-II, CV-III and CV-IV cultures were able to inhibit E. coli. While only CV-I showed antimicrobial activity against S. aureus with 0.4 cm of zone of inhibition. CV-I, CV-II showed inhibition of 0.7 and 0.9 cm respective against E. fecalis. CV-I and CV-IV showed good activity with 1.4 and 1.1 cm zone of inhibition against fungal pathogen C. albicans 3557. Antimicrobial activity is the important property for any probiotic micro-organism to prevent and cure enteropathogenic infections of hosts. Present study reported CV-I isolate to inhibit Gram positive as well as Gram negative pathogenic bacteria also possessing good activity against C.albicans, a commensal opportunistic pathogen in gastrointestinal tract of host.
Yeast Identification
Yeast isolate CV-I was genotypically sequenced and analyzed by ITS region. After comparing ITS gene region in NCBI database, the isolate CV-I was identified as Saccharomyces cerevisiae with ITS sequence as given below. AACTTAAAGGAATTGACGGAAGGGCACCACCAGGAGTGGAGCCTGCGGCTTAATTTGACTCAACACTGGGAAACTCACCAGGTCCAGACACCATAAGGATTGACAGATTGAGAGCTCTTTCTTGATTTTGTGGGTGGTGGTGCATGGCCGTTCTTCGTTGGTGGAGTGATTTGTCTGCTTAATTGCGATAACGAACGAGACCTTAACCTACTAAATAGTGGTGCTAGCATTTGCTGGTTATCCACTTCTTAGAGGGACTATCGGTTTCAAGCGGATGGAAGTTTGAGGCAATAACAGGTCTGTGATGCCCGTAGACGTTCTGGGCCGCACGCGCGCTACACTGACGGCGCCAGCGAGTCTAACCTTGGCCGAGAGGTCTTGGTAATCTTGTGAAACTCCGTGTGCTGGGGATAGAGCATTGTAATTATTGCTCTTCAACGAGGAATTCCTAGTAAGCGCAAGTCATCAGCTTGCGTTGATTACGTCCCTGCCCTTTGTACACACCGCCCGTCGCTAGTACAGATTGAATGGCTTAGTGAGGCCTCAGGATCTGCTTGGAGAAGGGGGCAACTCCATCTCAGAGCGGAGAATTGGGACAAACTTGGTCATTTAGAGGACCTAAAAGTCGTAACAAGGTTTCCGTAGGTGAACCTGCAGA
The isolate CV-I showed maximum similarity (98.18%) with Saccharomyces cerevisiae strain Makgeolli with Accession No. CP025108.1.
Caterpillars are very sensitive organisms and show dynamic nature in their gut microbiome diversity.16 Caterpillars lack prominent microbiome, as the diet shifts microbiome in gut of these organisms gets changed. However, there are some contrasting reports on correlation between diet and gut microbiome of caterpillar.17 Still there is an unknown role of structure and function of gut microbiome of caterpillar and yet have to be explored. This study reported cultures from frass that are proxy of gut microbiome of caterpillar (Indarbela quardinotata). The present report is the first of the kind in identification of Saccharomyces cerevisiae from frass of caterpillar and it’s evaluation as probiotic.
Conclusion
Due to the increased awareness of health food, probiotics are one of the most sorted out organisms. Many bacteria and yeast cultures are identified for their probiotic use in human and animals. Owing to the microbial diversity and functionality, still there is scope for isolation of microorganisms and screen them for probiotic potential. The present study is the first report of isolation of yeast from caterpillar frass and its probiotic characterization. Four yeast cultures isolated are non-hemolytic and have ability to grow at wide range of pH as well as in presence of intestinal environment. These cultures could inhibit common pathogenic organisms and shown good auto aggregation property. The best isolate, CV-I has been identified as Saccharomyces cerevisiae by molecular methods. Considering the results CV-I, is an excellent candidates for further probiotic characterization.
References
- Paraschiv D., Vasile A., Constantin M., Ciobanu A., Bahrim G. Study of Physiological properties of some probiotics in multiple cultures with mesophilic lactic acid bacteria by flora Danica CH. Hansen commercial starter. Food Technology. 2011;35(2):56-65.
- Charteris W. P., Kelly P. M., Morelli P., Collins J. K. Development and application of in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillusand Bifidobacterium species in upper human gastrointestinal tract. Journal of Applied Microbiology. 1998;84:759-768.
CrossRef - Yirga H. The use of probiotics in animal nutrition. Journal of probiotics & Health. 2015;3(2):1-10.
CrossRef - Chen L. S., Ma Y., Maubois J. L., He S. H., Chen L. J., Li H. M. Screening for the potential probiotic yeast strains from raw milk to assimilate cholesterol. Journal of Dairy Science Technology. 2010;90:537-548.
CrossRef - Engel P and Moran A. N. The gut microbiota of insects – diversity in structure and function. FEMS Microbiology Review. 2013;37:699-735.
CrossRef - Sonavale R. M., Kunchiraman B. N. Probiotic characteristics of anti-candida Bacillus tequilensis isolated from sheep milk and buffalo colostrums. International Journal of Health Sciences and Research. 2018;8(8):254-260.
- Lohith K and Anu Appaiah K. A. In vitro probiotic characterization of yeasts of food and environmental origin. International Journal of Probiotics and Prebiotics. 2014;9(3):87-92.
- Charteris W. P., Kelly P. M., Morelli P., Collins J. K. Development and application of in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in upper human gastrointestinal tract. Journal of Applied Microbiology. 1998;84:759-768.
CrossRef - Ogunremi O. R., Sanni A. I and Agrawal R. Probiotic potentials of yeasts isolated from some cereal-based Nigerian traditional fermented food products. Journal of Applied Microbiology. 2015;119:797-808.
CrossRef - Chelliah R., Ramakrishnan S. R., Prabhu P. R and Antony U. Evaluation of antimicrobial activity and probiotic properties of wild-strain Pichia kudriavzevii isolated from frozen idli batter. YEAST. 2016;33:385-401.
CrossRef - Palande V., Meora R., Sonavale R. M., Makashir M., Modak M. S., Kapse N., Dhakephalkar P. K., Ranjekar P. K., Kunchiraman B. N. Inhibition of pathogenic strains of Candida albicans and non-albicans by Bacillus species isolated from traditional Indian fermented food preparations. International Journal of Current Microbiology and Applied Sciences. 2015;4(3):691-699.
- Polkade A.V., Ramana V. V., Joshi A., Pardesi L and Shouche Y. S. Rufibacter immobilis nov., isolated from a high altitude saline lake. International Journal of Systemic and Evolutionary Microbiology. 2015;65;1592-1597.
CrossRef - Sourabh A., Kanwar S. S and Sharma O. P. In vitro characterization of Saccharomyces cerevisiae HM535662 obtained from an indigenous fermented food “Bhaturu” of Western Himalayas. African Journal of Biotechnology. 2012;11(52):11447-11454.
- Gupta A and Sharma N. Characterization of Potential Probiotic Lactic Acid Bacteria- Pediococcus acidilactici Ch-2 Isolated from Chuli- A Traditional Apricot Product of Himalayan Region for the Production of Novel Bioactive Compounds with Special Therapeutic Properties. Journal of Food: Microbiology, Safety & Hygiene. 2017;2(1):1-11.
- Shehata M. G., El Sohaimy S. A., El-Sahn A. M., Youssef M. M. Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Annals of agricultural science. 2016;61(1):65-75.
CrossRef - Gotcheva V., Hristozova E., Hristozova T., Guo M., Roshkova Z. &Angelov A. Assessment of potential probiotic properties of lactic acid bacteria and yeast strains. Food biotechnology. 2013;16(3):211-225.
CrossRef - Hammer T. J., Janzen D. H., Hallwachs D., Jaffe S. L., Fierer N. Caterpillar lack a resident gut microbiome. PNAS. 2017;114(36):9641-9646.
CrossRef - Chaturvedi S., Rego A., Lucas L. K., Gompert Z. Sources of variation in the gut microbial community of Lycaeides melissa caterpillars. Scientific Reports. 2017;7:1-13.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





