How to Cite | Publication History | PlumX Article Matrix
Yasser M. M , Mousa A. S. M
, Mousa A. S. M , Marym A. Marzouk
, Marym A. Marzouk and Tagyan A. I*
and Tagyan A. I*
Botany and Microbiology Department, Faculty of Science, Beni-Suef University, Egypt.
Corresponding Author E-mail: i_aya50@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2731
ABSTRACT: Solanum tuberosum L. possesses economic properties and can host endophytic mycoflora. A total of 19 endophytic fungi were identified via morphological and molecular methods. Among them, Trichoderma harzianum was the core-group fungus with a relative frequency of 36.7%. In the preliminary antimicrobial assay, all the test pathogens were inhibited by Alternaria tenuissima, Penicillium pinophilum and Penicillium rubens with a maximum inhibition zone of 26 mm and a minimum zone of 11 mm using agar-plug method. All the isolated endophytic fungi produced amylase, while cellulase and tyrosinase were recorded for most of the isolated species, whereas laccase and protease and manganese peroxidase were shown by a few taxa. None of the isolated fungi produced chitinase. This study revealed the biodiversity of endophytic fungi isolated from Solanum tuberosum that could be a promising source of bioactive compounds applied in many industries.
KEYWORDS: Biodiversity; Endophytic Fungi; Solanum tuberosum; Trichoderma harzianum
Download this article as:| Copy the following to cite this article: Yasser M. M, Mousa A. S. M, Marzouk M. A, Tagyan A. I. Molecular Identification, Extracellular Enzyme Production and Antimicrobial Activity of Endophytic Fungi Isolated from Solanum Tuberosum L. in Egypt. Biosci Biotech Res Asia 2019;16(1). |
| Copy the following to cite this URL: Yasser M. M, Mousa A. S. M, Marzouk M. A, Tagyan A. I. Molecular Identification, Extracellular Enzyme Production and Antimicrobial Activity of Endophytic Fungi Isolated from Solanum Tuberosum L. in Egypt. Biosci Biotech Res Asia 2019;16(1). Available from: https://bit.ly/2UuQ0mC |
Introduction
Solanum tuberosum L. (potato) belongs to the nightshade family (Solanaceae). The global output of potato in 2016 was amounted to be almost 376.83 million tons. The origin of the potato is the Andes of South America.1 It is common with major producers in China, Russia, Poland, the USA, Ukraine, Germany, and India. In 2017, the total exports of the Egyptian S. tuberosum were about 800,000 ton, so it is an important agricultural crop in Egypt.2
Endophytes are generally all microorganisms living within plants with no symptoms apparent to their hosts.3 The endophytic fungi hosted approximately one million species of plant.4 This group of fungi is ubiquitous.5 Recently, Trabelsi et al.6 isolated endophytic fungi from Solanum tuberosum such as Aspergillus flavus, Aspergillus niger, Penicillium chrysogenum, and Penicillium polonicum. Also, Marak and Kayang7 isolated a total of 44 endophytic fungi from Solanum tuberosum such as Aspergillus niger, Fusarium oxysporum, Penicillium funiculosum, Trichoderma harzianum.
Unlike, the utilized chemicals and pesticides for the disease overcoming, the endophytes were a cheap and environmentally safe source of bioactive compounds applied for protection against human pathogens.8 Nowadays, among the hardest troubles in the world were the developing of the non-susceptible drug in pathogenic microbes, such as Staphylococcus aureus that was resistant against methicillin.9 Deshmukh et al.10 have stated that endophytic fungi have antimicrobial compounds extracted from them. Endophytic fungi are a novel hope as various endophytic fungi have antifungal potential.11
Endophytic fungi were considered as a novel source for acquiring enzymes with unique prospects as their easy handling and cultivation, fast growth, and high yield.12 Various extracellular enzymes are secreted by endophytic fungi as cellulase, amylases, laccases, chitinases, and proteinases.13 Endophytic fungi have been investigated for several applications owing to their extracellular enzymes production.14 These enzymes operate many functions in an organism extending from obtaining nutrition from their host, food substances hydrolysis and are involved in eliciting defense mechanisms against pathogens. Subsequently, there is an urgent need to discover and use various unique enzymes with great stability for industrial purposes. Few studies have been done on endophytes from Solanum tuberosum in Egypt despite their importance. The present study aimed to isolate and identify endophytic fungi from Solanum tuberosum. Moreover, the isolated fungi were screened for their antimicrobial activity and extracellular enzymes production.
Materials and Methods
Location and Sampling
Solanum tuberosum L. (Potato) was collected from Beni-Suef governorate (latitude 29° 4′ 0″ N and longitude 31° 5′ 0″ E), Egypt during March 2015–March 2016. Plants with no visible symptoms of disease were selected and then transported to the laboratory in sterile bags and handled within 5 hours of sampling.
Surface Sterilization of Solanum Tuberosum and Isolation of Endophytic Fungi
According to the method of,15 plants were washed in running tap water to eliminate soil debris. Leaf, stem and root samples were cut into small segments of 1 cm long and 3 mm broad. Those segments were sunken in 70% ethanol for 1–3 minutes, 4% NaOCl for 1.5 minutes, 70% ethanol for 1minute and finally rinsed 3 times with sterile distilled water, then they were dried by sterile filter paper. Five segments were placed on potato dextrose agar medium with 250mg/l chloramphenicol. The plates were incubated at 25°C in a dark condition. The emerged fungi from segments were observed every 2 days for at least 3 weeks. The mycelia emerged from the segments were regularly isolated and the hyphal tips were transferred to the PDA plates free of antibiotics. Single spore cultures were prepared for each strain to ensure purity of strains.
Data Analysis
The colonization and relative frequency were calculated according to Petrini et al.16:
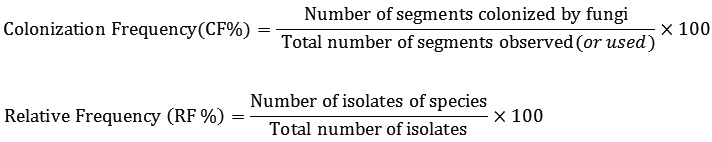
Morphological Identification of Endophytic Fungi
For morphological identification of endophytic fungi, hyphae and conidia were taken from purified colonies and examined by optical microscope according to.17
Molecular Identification of Endophytic Fungi
Genomic DNA was extracted from mycelia grown on 3% MEA(Malt Extract Agar) incubated at 28oC harvested after 2 days with the Plant DNeasy Minikit (QIAgen GmbH, Hilden, Germany) according to the manufacturer’s instructions. A region of nuclear DNA containing the ITS1 and two regions of the rRNA gene cluster was amplified by PCR using the primer combinations SR6R and LR1 (21) as described by Kullnig-Gradinger et al.18 PCR products were sequenced. The sequences were dropped in NCBI GenBank and compared with those available in the data base using a sequence similarity search tool (blastn). 19
Screening of Endophytic Fungi for Antimicrobial Activity
The preliminary screening of antimicrobial activity was carried out against Candida albicans, Salmonella typhi, Sarcina ventriculi and Staphylococcus aureus obtained from (Microbiology Laboratory, Faculty of Science, Ain Shams University, Egypt) using the agar plug method.20 The plates were poured with nutrient agar (NA) medium and inoculated with 100 µl of the pathogen suspension (1.5×108 CFU/ ml) then regularly spread by a sterile cotton swab on the medium. The grown mycelial discs (6 mm) of all isolates (15 day-olds) grown on potato dextrose agar (PDA) medium (potato infusion 200 g, dextrose 20 g, agar 20 g, distilled water 1 L and pH 6) were taken from actively growing edges of isolates using a sterile cork borer and put on the surface of nutrient agar medium (NA) (peptone 5g, yeast extract 5g, beef extract 3g ,agar 20g, distilled water 1 L and pH 7) previously inoculated with test organisms. The plates were wrapped by parafilm and incubated at 37°C for 24 h. Then, the antibacterial activity was documented by the visualization and estimation of inhibition zones.
Extracellular Enzyme Production
Amylase Production
The amylase production was screened using glucose yeast extract-peptone (GYP) agar medium (glucose 1g, yeast extract 0.1g, agar 15 g, distilled water 1 L and pH 6) which included 1% soluble starch. After incubation, the plates were flooded with 1% iodine and 2% potassium iodide. The clear zone surrounding the colony indicated amylase production.21
Cellulase Production
Cellulase production was estimated by GYP agar medium supplemented with 0.5% Na-carboxy-methylcellulose. Plates were incubated for 5 days at 28°C then immersed with a solution of 1% Congo red dye for 20 minutes and destained with 1N NaCl solution for 15 minutes. The cellulase enzyme was evaluated by the presence of a light yellow area around the colony of the fungus.21
Tyrosinase Production
Tyrosinase production was assessed using tyrosine agar medium (peptone 5g, beef extract 3g, agar 20g, L-tyrosine 5g and pH 7). The brown color around the colony indicated tyrosinase enzyme.9
Protease Production
Protease production was screened by GYP agar medium amended with 1% casein and pH 6.5 and incubated for 5 days. The clear zone around the colony indicated protease enzyme.21
Chitinase Production
Chitinase production was estimated by chitin agar medium (yeast extract 1.5 g, chitin 2.0 g, agar 20 g and distilled water 1 L). Plates were inoculated with test cultures and then incubated at 26°C up to 72 h. The appearance of a clear zone around the culture indicated the chitinase enzyme.
Estimation of Lignin-Degrading Enzymes
Manganese Peroxidase Production
Manganese peroxidase production was estimated using discs cut from the edge of 6 days old fungal cultures. The fungal discs were inoculated on Boyd and Kohlmeyer (B&K) medium (glucose 10 g, peptone 2 g, yeast extract 1 g, agar 18 g, distilled water 1 L and pH 6.0) amended with 4 mM guaiacol.22 The plates were incubated at 25±2°C for 2 weeks after enclosing them with black polythene bags. The formation of reddish brown color under and around the fungal colony was owing to guaiacol oxidation.
Laccase Production
Laccase production was performed using GYP agar medium supplemented with 0.005% 1-naphthol. The blue color indicated laccase enzyme due to oxidation of 1-naphthol.23
Statistical Analysis
The tests were performed in triplicate and the results were analyzed statistically by the SPSS program version 20. The analyses of variance were according to the rules of the ANOVA. The significance of differences between the means as determined through Duncan’s multiple range Test.
Results
Isolation and Identification of Endophytic Fungi
In the present study, 2520 potato segments analyzed (840 of leaves, 840 of stems and 840 of roots segments), 1128 isolates were taxonomically identified. Colonization frequencies of endophytic fungi from leaves, stems, and roots were 50.9, 41.9 and 41.4%, respectively. The percentages of endophytic fungi isolated from leaves, stems, and roots were 37.9, 31.2, and 30.9%, respectively. Morphological and molecular identification using ITS revealed nineteen taxa of endophytic fungi (Figure 1 & Table 1). All the identified endophytic fungi belong to Ascomycota. The isolates belonging to three classes, Sordariomycetes, Dothideomycetes, and Eurotiomycetes. Trichoderma harzianum was the most dominant endophytic fungus with a relative frequency (RF) of 36.7%, Aspergillus niger was the second most one with RF=22.2% while Penicillium pinophilum and Penicillium polonicum showed the minimum percentage (RF=0.12%) for each (Table1). On the other hand, the genus Penicillium was the most diverse (5 different species) among other taxa. All isolates showed a good similarity (98 – 100 %) to the GenBank sequences (Table 1).
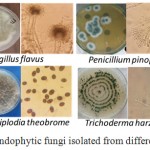 |
Figure 1: Pure cultures of some endophytic fungi isolated from different parts of the potato plant. |
Table 1: List of endophytic fungi isolated from different parts of potato and their GenBank accession numbers and relative frequency.
| TUCIM No. | Most similar GenBank entry | Species identification | % similarity / % query coverage | Number of isolated colonies | % Relative frequency | |||
| Root | Stem | Leaf | Total | |||||
| 6640 | KX664322 | Alternaria tenuissima | 99/99 | 12 | 69 | 8 | 89 | 7.9 |
| 6676 | JF951750 | Aspergillus flavus | 99/90 | 27 | 63 | 10 | 100 | 8.9 |
| 6670 | KF154412 | Aspergillus niger | 100/96 | 62 | 69 | 119 | 250 | 22.2 |
| 6644 | KC859010 | Aspergillus ochraceus | 100/93 | _ | 4 | 9 | 13 | 1.15 |
| 6671 | KP307914 | Aspergillus oryzae | 99/99 | 13 | 2 | _ | 15 | 1.2 |
| 6637 | NR_144848 | Chaetomium cervicicola | 98/100 | _ | 4 | _ | 4 | 0.35 |
| 6645 | KF753941 | Curvularia lunata | 100/96 | 23 | _ | 10 | 33 | 2.9 |
| 6636 | KC311517 | Fusarium equiseti | 99/99 | 12 | 5 | _ | 17 | 1.5 |
| 6634 | HF546381 | Fusarium nygamai | 99/99 | _ | 2 | 39 | 41 | 3.6 |
| 6635 | LT746253 | Fusarium oxysporum | 99/99 | _ | _ | 3 | 3 | 0.27 |
| 6649 | KY655194 | Lasiodiplodia theobrome | 100/96 | _ | _ | 4 | 4 | 0.4 |
| 6641 | GU183120 | Penicillium funiculosum | 99/98 | _ | 14 | 22 | 36 | 3.2 |
| 6642 | JN620402 | Penicillium minioluteum | 99/94 | _ | _ | 40 | 40 | 3.5 |
| 6668 | AB606412 | Penicillium pinophilum | 99/98 | _ | _ | 2 | 2 | 0.12 |
| 6669 | KX958077 | Penicillium polonicum | 99/99 | _ | 2 | _ | 2 | 0.12 |
| 6673 | LC105692 | Penicillium rubens | 100/97 | _ | _ | 18 | 18 | 1.6 |
| 6648 | MG065799 | Stemphylium vesicarium | 99/100 | 14 | _ | _ | 14 | 1.24 |
| 6638 | KM491893 | Trichoderma harzianum | 99/99 | 175 | 102 | 137 | 414 | 36.7 |
| 6647 | KU945936 | Ulocladium sp. | 100/96 | 10 | 16 | 7 | 33 | 2 |
| _ | Total | _ | 348 | 352 | 428 | 1128 | _ | |
Preliminary Screening of Antimicrobial Activity
Out of the 19 isolates, three isolates were able to inhibit all the test organisms. The results revealed that all the endophytic fungi exhibited a varied degree of inhibition against the test pathogens (Figure 2). Out of the 19 isolates only 10, endophytic fungi revealed inhibition of at least one of the test pathogens. Candida albicans and Salmonella typhi were inhibited by seven strains while Sarcina ventriculi and Staphylococcus aureus were inhibited by eight and four, respectively (Figure 2 & Table 2). Among the tested endophytic fungi, Alternaria tenuissima Penicillium pinophilum and Penicillium rubens inhibited all the test pathogens with a maximum zone of inhibition of 26 mm and a minimum of 11 against test pathogens, while all the other endophytic fungi except Alternaria tenuissima, Aspergillus flavus, Fusarium equiseti, Fusarium oxysporum, Lasiodiplodia theobromae, Penicillium funiculosum, Penicillium pinophilum Penicillium rubens, Stemphylium vesicarium, and Ulocladium sp. showed no inhibition to test pathogens (Table 3).
 |
Figure 2: Antimicrobial activity of six endophytic fungi against human pathogens (a, b, c and d) positive results while (e and f) negative results,
|
a- Alternaria tenuissima against Candida albicans, b- Penicillium pinophilum against Salmonella typhimurium, c- Fusarium oxysporum against Sarcina ventriculi, d- Penicillium pinophilum against Staphylococcus aureus, e- Curvularia lunata against Sarcina ventriculi and f- Fusarium nygamai against Candida albicans.
Table 2: Preliminary screening of antimicrobial activity of cultures of endophytic fungi isolated from different parts of the potato plant.
| Endophytic fungi | Zone of inhibition (mm) | |||
| Candida albicans | Salmonella typhimurium | Sarcina ventriculi | Staphylococcus aureus | |
| Alternaria tenuissima | 26.0±0.35g | 21.0±0.3e | 20.0±0.47d | 11.0±0.25b |
| Aspergillus flavus | 19.0±0.35d | 11.0±0.3a | 15.0±0.47b | – |
| Aspergillus niger | – | – | – | – |
| Aspergillus ochraceus | – | – | – | – |
| Aspergillus oryzae | – | – | – | – |
| Chaetomium cervicicola | – | – | – | – |
| Curvularia lunata | – | – | – | – |
| Fusarium equiseti | 20.0±0.35e | 23.0±0.3f | 21.0±0.47e | – |
| Fusarium nygamai | – | – | – | – |
| Fusarium oxysporum | – | 13.0±0.3b | 23.0±0.47g | – |
| Lasiodiplodia theobromae | – | – | – | 10.0±0.25a |
| Penicillium funiculosum | 16.0±0.35c | 19.0±0.3d | 11.0±0.47a | – |
| Penicillium minioluteum | – | – | – | – |
| Penicillium pinophilum | 25.0±0.35f | 25.0±0.3g | 22±0.47f | 18.0±0.25d |
| Penicillium polonicum | – | – | – | – |
| Penicillium rubens | 11.0±0.35a | 15.0±.3c | 17.0±0.47c | 13.0±0.25c |
| Stemphylium vesicarium | – | – | 20.0±0.47d | – |
| Trichoderma harzianum | – | – | – | – |
| Ulocladium sp. | 13.0±0.35b | _- | _- | -_ |
Values are means of three independent replicates. ± indicate standard error. Means followed by the same letter within the same column are not significantly different according to Duncan test (P ≤ 0.05).
Enzyme Production by Endophytic Fungi
All the 19 fungal isolates secreted amylase. Cellulase was secreted by twelve while tyrosinase, protease, manganese peroxidase, and laccase were produced by six, three, eleven and two, respectively. None of the isolated fungi produced chitinase (Table 3 and Figure 3).
Table 3: Extracellular enzyme production of endophytic fungi isolated from different parts of the potato plant.
| Endophytic fungi | Amylase | Cellulase | Tyrosinase | Protease | Chitinase | Manganese peroxidase | Laccase |
| Alternaria tenuissima | +++ | – | ++ | – | – | – | +++ |
| Aspergillus flavus | +++ | +++ | +++ | – | – | – | – |
| Aspergillus niger | + | +++ | – | – | – | – | – |
| Aspergillus ochraceus | + | ++ | – | + | – | – | – |
| Aspergillus oryzae | + | + | +++ | +++ | – | – | – |
| Chaetomium cervicicola | + | ++ | +++ | – | – | +++ | – |
| Curvularia lunata | ++ | + | +++ | – | – | + | + |
| Fusarium equiseti | ++ | – | – | – | – | – | – |
| Fusarium nygamai | ++ | ++ | +++ | – | – | – | – |
| Fusarium oxysporum | ++ | – | +++ | – | – | – | – |
| Lasiodiplodia theobromae | +++ | +++ | – | – | – | – | – |
| Penicillium funiculosum | + | + | ++ | – | – | – | – |
| Penicillium minioluteum | + | + | – | – | – | – | – |
| Penicillium pinophilum | + | – | – | – | – | + | – |
| Penicillium polonicum | + | +++ | +++ | – | – | – | – |
| Penicillium rubens | + | + | – | – | – | +++ | – |
| Stemphylium vesicarium | ++ | – | ++ | – | – | – | – |
| Trichoderma harzianum | +++ | – | – | – | – | +++ | – |
| Ulocladium sp. | ++ | – | ++ | ++ | – | + | – |
Where- = No production; + = Weak production; ++ = Medium production; +++ = High production.
 |
Figure 3: Enzyme production by endophytic fungi isolated from different parts of potato plant, (a and b) amylase production, (c and d) cellulase production, (e and f) tyrosinase production, (g and h) protease production, (i and j) manganese peroxidase production, (k and l) laccase production.
|
Discussion
The results showed that higher number endophytic fungi were isolated from leaves than stems and roots tissues. This may be attributed to the large surface area of leaves exposed to the surroundings and the presence of stomata that aid fungal mycelium entrance.24 The morphological and molecular identification of isolated endophytic fungi revealed 19 species. It was believed that one plant can be a habitat of six fungi, but after adding fungal endophytes, the ratio of fungal: plant species has now been altered to 33:1.25 The low rate of colonization (19 species) may be ascribed to the secretion of the phytochemicals.26
Aspergillus, Fusarium, and Penicillium were the predominant genera. Their prevalence may owe to the high spore production that facilitates their widespread and getting established as endophytes.27
In the preliminary antimicrobial assay, three strains showed inhibition to all the test pathogens (Candida albicans, Salmonella typhi, Sarcina ventriculi and Staphylococcus aureus). Fusarium equiseti and Penicillium pinophilum revealed significant inhibition of Salmonella typhimurium. This may drive to find a new antibiotic against typhoid caused by this bacteria.
Fungal amylase has enormous uses in the food and pharmaceutical industries.14 In this study, all endophytes degrade starch by amylase enzyme. Out of the 19 endophytic isolates only 12, endophytic fungi produced cellulase enzyme and this enzyme involved in biofuels production.28 It was established that Ulocladium sp., Aspergillus oryzae, and Aspergillus ochraceus produced protease enzyme, almost this enzyme is applied in treatments of diabetes.29 Tyrosinase was produced by eleven strains and it is known for lignin degradation and synthesis of melanin.14 Alternaria tenuissima and Curvularia lunata showed laccase production. Laccase plays a role in the degradation of lignin, and can, therefore, be classed as lignin-modifying enzymes.30 Lately, the effectiveness of laccases has also been applied to nanobiotechnology.31 In similar studies, Rajagopal et al.26 reported that Curvularia lunata produced laccase. Among all isolates, only six isolates produced manganese peroxidase and this enzyme used for the degradation of lignin that plays a major role in biomass biodegradation.32 Endophytic fungi produced a variety of enzymes in this study that may work as a unique source for applications in industry.
Conclusion
In the present study, 19 endophytic fungi were isolated and identified from leaves, stems, and roots of the potato plant. These fungi were able to produce extracellular enzymes such as amylase, cellulase, tyrosinase, protease, manganese peroxidase and laccase. Also, Fouda et al.13 were able to produce enzymes such as cellulase, amylases, laccases, chitinases, and proteinases from endophytic fungi. The results demonstrated that all the test pathogens were inhibited by Alternaria tenuissima, Penicillium pinophilum and Penicillium rubens. This evidenced that endophytic fungi have antimicrobial compounds extracted from them Deshmukh et al.10 According to the obtainable results, it can be obviously realized that endophytes isolated from potato plants could be valuable and applicable in many industries, mainly the pharmaceutical industries.
Acknowledgments
The research was funded by the Faculty of Science, Beni-Suef University, Egypt. Authors thank Marica Grujic, Microbiology, and Applied Genomics Group, Institute of Chemical, Environmental & Bioscience Engineering Vienna University of Technology, Austria for her help during the molecular identification of isolates.
Conflict of Interest
There is no conflict of interest.
Reference
- Spotorno A. E & Veloso A. Flora and fauna. In The Aymara Springer, Dordrecht.starch-converting enzymes of the α-amylase family. J. Biotechnolm. 1990;94:137–155. 19-32.
CrossRef - Ahmed A. I. S.Biological Control of Potato Brown Leaf Spot Disease Caused by Alternaria alternata Using Brevibacillus formosus Strain DSM 9885 and Brevibacillus brevis Strain NBRC 15304. J Plant Pathol Microbiol. 2017;8(413):2.
CrossRef - Azevedo J. L., Maccheroni Jr W., Pereira J. O & de Araújo W. L. Endophytic microorganisms: a review on insect control and recent advances on tropical plants. ELECTRON J BIOTECHN. 2000;3(1):15-6.
CrossRef - Gurupavithra S & Jayachitra A. Isolation and identification of endophytic fungi from Ocimum sanctum and analyse its antioxidant properties. IJMRPS. 2013;4(4):1120-1125.
CrossRef - Zabalgogeazcoa I. Fungal endophytes and their interaction with plant pathogens: a review. SPAN J AGRIC RES. 2008;6(S1):138-46.
CrossRef - Trabelsi B. M., Abdallah R. A. B., Ammar N., Kthiri Z & Hamada W. Bio-suppression of Fusarium Wilt Disease in Potato Using Nonpathogenic Potato-associated Fungi. J Plant Pathol Microbiol. 2016;7(347):2.
- Marak M. C. N & Kayang H. Isolation and Identification of Endophytic Fungi Associated with Solanum tuberosum L. of South-West Garo Hills, Meghalaya. IJAAST. 2018;5(1):58-65.
- Talapatra K., Das A. R., Saha A. K & Das P. In vitro antagonistic activity of a root endophytic fungus towards plant pathogenic fungi. JABB. 2017:5(02):068-71.
- Majumdar D. R., Singh R., Dondilkar S., Shaikh N., Pawale G., Shinde P & Sakate P. Enzyme array from thermophilic fungal isolate RSND. JPR. 2018;7(4):60-70.
- Deshmukh S. K., Verekar S. A & Bhave S. V. Endophytic fungi: a reservoir of antibacterials. FRONT MICROBIOL. 2015;5:715.
CrossRef - Strobel G & Daisy B. Bioprospecting for microbial endophytes and their natural products. MICROBIOL MOL BIOL R. 2003;67(4):491-502.
CrossRef - Carrim A. J. I., Barbosa E. C & Vieira J. D. G. Enzymatic activity of endophytic bacterial isolates of Jacaranda decurrens Cham.(Carobinha-do-campo). BRAZ ARCH BIOL TECHN. 2006;49(3):353-9.
CrossRef - Fouda A. H., Hassan S. E. D., Eid A. M & Ewais E. E. D. Biotechnological applications of fungal endophytes associated with medicinal plant Asclepias sinaica (Bioss.). AOAS. 2015;60(1):95-104.
CrossRef - Pavithra N., Sathish L & Ananda K. Antimicrobial and enzyme activity of endophytic fungi isolated from Tulsi. JPBMS. 2012;16(16):2014.
- Arnold A. E., Maynard Z & Gilbert G. S. Fungal endophytes in neotropical trees: abundance, diversity, and ecological implications. Tropical ecosystems: structure, diversity and human welfare. New Delhi: Oxford & IBH. 2001;739-43.
- Petrini O., Stone J & Carroll F. E. Endophytic fungi in evergreen shrubs in western Oregon: a preliminary study. Canadian Journal of Botany. 1982;60(6):789-96.
CrossRef - Moubasher A. H. Soil fungi in Qatar and other Arab countries. The Centre for Scientific and Applied Research, University of Qatar, Doha, Qatar. 1993;568.
- Kullnig-Gradinger C. M., Szakacs G & Kubicek C. P. Phylogeny and evolution of the genus Trichoderma: a multigene approach. Mycological Research. 2002;106(7):757-67.
CrossRef - Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J MOL BIOL. 1990;215(3):403-10.
CrossRef - Zhang Y., Mu J., Feng Y., Kang Y., Zhang J., Gu P. J & Zhu Y. H. Broad-spectrum antimicrobial epiphytic and endophytic fungi from marine organisms: isolation, bioassay and taxonomy. Marine drugs. 2009;7(2):97-112.
CrossRef - Rajput K., Chanyal S & Agrawal P. K. Optimization of protease production by endophytic fungus, Alternaria alternata isolated from gymnosperm tree- Cupressus torulosa Don. W JPPS. 2016;5(7):1034-1054.
- Atalla M. M., Zeinab H. K., Eman R. H., Amani A. Y & Abeer A. A. E. A. Screening of some marine-derived fungal isolates for lignin degrading enzymes (LDEs) production. ABJNA. 2010;1(4):591-9.
- Sathish L., Pavithra N & Ananda K. Antimicrobial activity and biodegrading enzymes of endophytic fungi from eucalyptus. Int J Pharm Sci Res. 2012;3(8):2574-2583.
- Gond J. P., Grubnic S., Herzig C & Moon J. Configuring management control systems: Theorizing the integration of strategy and sustainability. AMJ. 2012;23(3):205-223.
CrossRef - Hawksworth D. L & Rossman A. Y. Where are all the undescribed fungi?. Phytopathology. 1997;87(9):888-891.
CrossRef - Rajagopal K., Meenashree B., Binika D., Joshila D., Tulsi P. S., Arulmathi R & Tuwar A. My codiversity and biotechnological potential of endophytic fungi isolated from hydrophytes. Curr Res Environ Appl Mycol J Fungal Biol. 2018;8(2):172-82.
CrossRef - Uzma F., Konappa N. M & Chowdappa S. Diversity and extracellular enzyme activities of fungal endophytes isolated from medicinal plants of Western Ghats, Karnataka. EJBAS. 2016;3(4):335-342.
CrossRef - Aguiar A & Ferraz A. Mecanismos envolvidos na biodegradação de materiais lignocelulósicos e aplicações tecnológicas correlatas. Química Nova. 2011;34(10):1729-1738.
CrossRef - Hua Y & Nair S. Proteases in cardiometabolic diseases: pathophysiology, molecular mechanisms, and clinical applications. Biochim Biophys Acta Mol Basis Dis. 2015;1852(2):195-208.
CrossRef - Cohen R., Persky L & Hadar Y. Biotechnological applications and potential of wood-degrading mushrooms of the genus Pleurotus. Microbiol. Biotechnol. 2002;58(5):582-94.
CrossRef - Kunamneni A., Plou F. J., Ballesteros A & Alcalde M. Laccases and their applications: a patent review. Recent Pat Biotechnol. 2008;2(1):10-24.
CrossRef - Knežević A., Milovanović I., Stajić M., Lončar N., Brčeski I., Vukojević J & Ćilerdžić J. Lignin degradation by selected fungal species. Bioresource Technology. 2013;138:117-23.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





