How to Cite | Publication History | PlumX Article Matrix
Muhammad Sabir1, Yasir Anwar2, Akram Khan1, Muhammad Ali1, Peerzada Yasir Yousuf3, Khalid Al-Ghamdi2 and Khalid Rehman Hakeem*2
1Institute of Biotechnology and genetic Engineering, Faculty of Crop production science, KPK Agriculture University, Peshawar Pakistan.
2Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia.
3Molecular Ecology Laboratory, Department of Botany, Faculty of Science, Jamia Hamdard, Hamdard Nagar New Delhi-110062, India.
Corresponding Author E-amil: kur.hakeem@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2750
ABSTRACT: Potato (Solanum tuberosum), one of the indispensable food crops, is susceptible to various fungal phyto-pathogenic infections that result in considerable production losses both in terms of quality and quantity. Developing fungal-resistant cultivars by introducing pathogen-resistant genes through transgenic approach has been a powerful tool to provide defense against the fungal pathogens. The current study was undertaken to develop a fungal resistant trait in a local potato variety. To achieve this goal, a local Pakistani Potato variety (Diamant), was transformed with chitinase ChiC gene utilizing plasmid pEKB/ChiC using Agrobacterium tumefaciens strain EHA101. The infected explants were grown on MS medium supplemented with 2 mg/l BAP and 0.2 mg/l NAA. Explants were then sub-cultured on MS medium supplemented with 2mg/l BAP and 2mg/l GA3 for shooting. PCR examination confirmed the integration of ChiC gene in the potato genomic DNA. The transformed potato variety could also be used as fungus-resistant breeding material and offers new opportunities to develop improved potato cultivars for different agronomic and other desirable traits.
KEYWORDS: ChiC; Chitinase; Diamant Potato; Transformation; Transgenic Plants
Download this article as:| Copy the following to cite this article: Sabir M, Anwar Y, Khan A, Ali M, Yousuf P. Y, Al-Ghamdi K, Hakeem K. R. ChiC Gene Enhances Fungal Resistance in Indigenous Potato Variety (Diamant) Via Agrobacterium-Mediated Transformation. Biosci Biotech Res Asia 2019;16(2). |
| Copy the following to cite this URL: Sabir M, Anwar Y, Khan A, Ali M, Yousuf P. Y, Al-Ghamdi K, Hakeem K. R. ChiC Gene Enhances Fungal Resistance in Indigenous Potato Variety (Diamant) Via Agrobacterium-Mediated Transformation. Biosci Biotech Res Asia 2019;16(2). Available from: https://bit.ly/2XTDU8q |
Introduction
Production of potato (Solanum tuberosum L), one of the mankind’s most valuable food crops [1] is seriously constrained due to occurrence of number of diseases. Fungi, bacteria, virus and viroids are the main disease-causing organisms which bring significant losses in potato production both in terms of quality and quantity[2]. Fungi in particular, cause more than 25 diseases including black dot, charcoal rot, brown spot, common rust, deforming rust, early blight, fusarium dry rot, leak, gangrene, etc in potato plants limiting the production to a large extent. It is thus need of the hour to manage fungal diseases in potato plants in order to sustain their productivity. Although fungicides are used for the control of fungal diseases but their adverse effects on the environment do not encourage their use [3],[4]. Genetic engineering technology is a powerful tool for the introduction of agronomically desired traits into commercial cultivars and is environment-friendly. However, for the development of transgenic plants, identification of suitable target genes and protocols for efficient gene transfer are essential [5]. With the advent of omics approaches, number of genes (encoding pathogenesis-related proteins) have been identified which can be manipulated to develop fungus-resistant plants. The best characterized pathogenesis-related genes are those encoding chitinase, a lytic enzyme that provides defense to plants specifically against fungal phyto-pathogens[6]. Chitinases catalyze the hydrolytic cleavage of the β-1, 4-glycoside bond of chitin, a key component of fungal cell wall [7]. The resulted disruption of the cell wall makes the fungal cells osmotically unstable and malformed by affecting the morphology and growth of fungal cells that leads to their death. Chitinases (E.C 3.2.2.14), the “glycosyl hydrolases” whose molecular weight ranges from 20 kDa to around 90 kDa [8], thus exhibit enormous potentiality to control the pathogenic plant fungi.
The presence of Chitinases varies from micro-organisms (e.g. Bacteria, actinomycetes, Yeasts, Fungi) to macro-organisms (e.g. arthropods, plants and humans). Bacteria such as Streptomyces [9], E. coli [10], Alteromonas [11] and Aeromonas [12] have the ability to produce Chitnases and such bacteria has been isolated from many sources including shellfish waste, soil, compost wastes of garden and parks and hot-springs. Chitinase has received high attention for bio-control of harmful insects and fungal phytopathogens. The introduction and expression of chitinase gene from different origins have resulted in the enhanced fungal resistance in different crops including tomato [13], Rice [14-15], Groundnut and Peanut [16,17,18], Banana [19], Italian rye grass [20], Cucumber [21], Cotton [22], Banana [23], Grapevine [24] and Tobacco [25]
In the current study, Diamant, a commercial potato variety of Pakistan was transformed with chitinase gene, Chi, to increase its resistant against fungal phyto-pathogens and sustain its productivity.
Materials and Method
Plant Material
Fresh tubers of Diamant (a commercial potato cultivar), were collected from National Agriculture Research Centre (NARC), Islamabad. These tubers after authenticated by a Taxonomist from University of Agriculture, Peshawar, were planted in pots in green house and were watered on a regular basis until 20 days. When the plants were completely developed, shoots and leaves were taken for pre-culturing. Leaves were washed several times with tap water and then surface-sterilized with sodium hypochlorite solution (1% active chlorine with 2-3 drops of Tween 20) for 20–25 min. The leaves were rinsed with sterilized water four times inside laminar flow cabinet. Leaf discs were then prepared with a sterilized scalpel (5 × 5 mm2) and used for pre-culture purposes. The experiments were carried out at Recombinant DNA Technology Laboratory, Institute of Biotechnology and Genetic Engineering (IBGE), Agricultural University Peshawar.
Bacterial strain and Transformation
A colony of Agrobacterium tumefaciens strain EHA101 pEKB containing Chitinase was cultured in LB medium in a shaker incubator at 180 rpm and 28 °C to attain OD of 0.8 at 600 nm. The bacterial suspensions were grown overnight, centrifuged and then prepared for inoculation.
Plant Transformation and Selection
Agrobacterium strain EHA101 harboring the binary vector plasmid pEKB/ChiC was used to infect the explants. MS medium lacking agar was prepared in two separate 100 ml flasks and autoclaved at a pressure of 15 psi and temperature of 121°C for 20 min. Agrobacterium tumefaciens was cultured overnight on a rotary shaker (130 rpm) at 28°C in liquid LB (Lysogeny Broth) medium (containing 10 g l-1 NaCl, 5 g l-1 yeast extract, 10 g l-1 tryptone, 25 mg l-1 chloramphenicol, 50 mg l-1 kanamycin sulphate with pH 7.2). The bacterial suspension was centrifuged at room temperature for 15 min at 8000 rpm in 15 ml falcon tubes. Supernatant was discarded and the bacterial pellet was resuspended in 100 ml flask containing hormone-free MS medium. The resulting Agrobacterium suspension (inoculum) was used for infection of the explants. All the explants were added to the inoculum in the flasks and were shaken continuously for 8–10 min. The leaves discs were blotted dry with sterilized filter paper to remove extra bacterial culture.
Cultivation on MS Media
The explants were then co-cultivated on 1% agar-containing solidified MS medium supplemented with 2 mg l-1 benzyl aminopurine (BAP), 0.2 mg l-1 1-naphthalene acetic acid (NAA) and 20 g l-1 sucrose, under continuous dim light using cool white fluorescent lamps for 3 days. The explants which were not infected with Agrobacterium and served as control were also treated likewise.
Co-cultivation
After co-cultivation, the explants were washed with hormone-free liquid MS medium containing 200 mg l-1 cefotaxime and transferred to agar-solidified MS medium supplemented with 200 mg l-1 cefotaxime as bactericide and 50 mg l-1 kanamycin sulphate (Km) as selective chemical. These antibiotics were added after filter-sterilizing and autoclaving. The cultured explants were kept in normal-culture room for 2 weeks. These explants were transferred to MS medium containing 2 mg l-1gibberellic acid (GA3), 2 mg l-1 Benzyl aminopurine (BAP), 1% agar (w/v), and 200 mg l-1 cefotaxime.
PCR Analysis
For molecular analysis, DNA was extracted from the young leaves of transgenic and non-transgenic potato plants using CTAB method. The forward (ChiC1- 5′-CGGGATCCGTCATGAGTCTGCTGGTCGC-3′) and reverse (ChiC2- 5′-ACGCGTCGACATCAGCAGCTCAGGTTCGGAC-3) primers of the chitinase gene were designed and used for amplification of ChiC gene in the Agrobacterium plasmid by PCR. The anticipated PCR product size was around 0.9 kb of chitinase gene. Electrophoresis of the PCR products was carried out using 1% agarose gel.
Results
Potato Transformation
Competent cells
The competent cells were prepared using calcium chloride Method [26] followed by transformation of E. coli with vector plasmid pEKB/ChiC (Fig. 1 and Fig 4 A), harboring ChiC gene, isolated from Streptomyces griseus strain HUT6037. Transformation of E. coli was carried out employing the method of Sambrook and Russell (2001) [27] that is shown in the (Fig. 2). The plasmid extracted from the E. coli was subjected to single and double digestion using Bam H I and Hind III for confirmation. Gel electrophoresis of the digested product was carried out and observed as shown in the Fig. 4B.
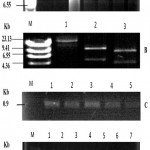 |
Figure 1: T- pEKH/ChiC DNA region having bar and chitinase genes.
|
The bar and chitinase genes are driven by CaMV 35S promoter, and for neomycin phosphotransferase (NPTII) gene by nopaline synthase promoter (nos-p). RB indicates right border and LB the left border sequences of the T-DNA region. CaMV 35S-P signifies Cauliflower mosaic virus 35S promoter and nos-T the terminator of the nopaline synthase gene. Restriction enzymes recognition sites are also indicated.
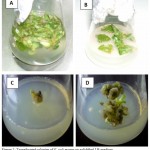 |
Figure 2: Transformed colonies of E. coli grown on solidified LB medium, containing 50 mgl-1 Kanamycin, and 20 mgl-1 Teramycin.
|
Potato Transfection
The Agrobacterium tumefaciens-mediated transformation of potato ‘Diamant’ with ChiC gene was done as shown in Fig. 3. Inoculum preparation was carried out by culturing Agrobacterium tumefaciens EHA101 over night. The leaves of explants were infected and co-cultivated. Callus formation was induced on medium having 50 mg/l kanamycin and 300 mg/l cefotaxime after 4 weeks of transfection. After 7 weeks of infection, shoot were generated on medium having 100 mg/l kanamycin and 300 mg/l cefotaxime (Fig. 3A, B, C and D). Similar results was also found using Agrobacterium tumefaciens-mediated transformation in shepody and favorite potato varities with 67 % of efficiency [28]. Another study shows that ODREB2B gene is transform into zhongshu and huangmazi potato varieties using agrobacterium tumefaciens [29].
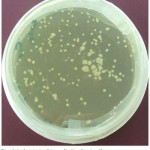 |
Figure 3: Agrobacterium tumefaciens-meditaed transformation with ChiC gene of potato ‘Diamant’.
|
(A) Leaf explants in the course of infection. (B) Explants in the course of co-cultivation. (C) Callus formation in infected explants after 4 weeks infection, grown on 50 mg/l kanamycin and 300 mg/l cefotaxime. (D) Shoot production in explants after 7 weeks infection, grown on100 mg/l kanamycin and 300 mg/l cefotaxime.
Molecular Analysis
For the screening of transformants for ChiC genes integration, genomic DNA was extracted using Cetyl Trimethyl Ammonium Bromide (CTAB) method [30] from the calli developed on MS medium containing kanamycin. Amplified fragments of approximately 0.90 kb were observed for all clones of ChiC suggesting the successful integration of transgene into the genome of transformed plants (Fig. 4C and 4D). PCR products from total DNA of non-transformed control plants showed no such amplified band.
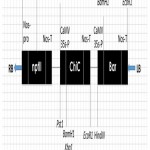 |
Figure 4: (A) Plasmid DNA extracted from the E.coli colonies grown on supplemented with 50 mg/l kanamycin supplemented LB medium.
|
M is size marker (λ/Hind III digest). 1-7 lanes illustrate plasmid DNA extracted from independent E. coli colonies. (B) Plasmid DNA extracted from E .coli by restriction enzyme digestion grown on 50 mg/l kanamycin supplemented LB media. M, size marker (λ/Hind III digest). Lane 1 shows uncut plasmid (23 kb). Lane 2 shows two fragments of the plasmid digested by Hind III. Lane 3 shows plasmid DNA double digestion with Hind III and Bam HI. (C) Agrobacterium plasmid DNA PCR Analysis. PCR analysis of plasmid DNA extracted from the Agrobacterium tumefaciens colonies grown on LB medium by using the ChiC gene primer. M is size marker (λ/Hind III digest). 1-5 lanes shows independent colonies in which ChiC gene is amplified (0.9 Kb). (D) PCR analysis of dna extracted from the leaf explant calli infected with the A. tumefaciens having ChiC gene, after 40 days of infection. M is size marker (1 Kb ladder digest). 1-5 lanes shows independent samples of explant in which ChiC gene was amplified (0.9 Kb). Lane 6 shows DNA from non-transformed control plants as a negative control. Lane 7, plasmid DNA from transformed Agrobacterium tumefaciens, as a positive control (the plasmid DNA).
Discussion
Fungal phyto-pathogenic infections are one of the major constraints to agricultural production. Almost 25% of plant productivity in developed countries and 50% in developing countries is lost due to plant diseases of which one third is only due to fungal infections [31]. Chemical fungicides are being applied to the plants to encounter the fungal infections but their use is associated with environmental risks. One of the advanced and eco-friendly solution to this conundrum is the development of fungal-resistant plants though modern biotechnological approaches. Genes which provide defense against the pathogen infections have been identified as pathogen-related genes (PR genes). Chitinase genes are one of the important PR genes that exhibit a broad-spectrum antifungal activity by catalyzing hydrolysis of chitin, a key component of fungal cell wall. In addition, these genes have also been reported to play vital role in abiotic stress tolerance of plants as reviewed by [32]. Chitinase genes have been used to produce genetically modified (GM) plants that are resistant to fungi. Cloning, expression and introduction of the chitinase gene into biologically-susceptible species have been one of the appealing areas of chitinase applications. The Chitinase genes have been characterized from different microorganisms [33],[34],[35],[36] and transformed into plants and bacterial strains to enhance their antifungal activity[37].
Introducing chitinase genes in susceptible plants have been reported to induce enhanced protection by degrading the chitin in hyphae restricting fungal growth, and by inducing the release pathogen-borne elicitors that augment defense reaction. The expression of chitinase gene has been observed to induce resistance to fungal infections in various transgenic crops including wheat [38], rice [39], tomato [13], cotton [40], apple [41] and banana [15] etc. In this study, Diamant, a commercial potato cultivar was transformed with ChiC gene to increase its tolerance against fungal phyto-pathogenic infections. This is the first report of Agrobacterium mediated transformation with Chitinase gene in Pakistani commercial potato variety, Diamant. The lower transformation efficiency observed in our study may be due to hormonal combination, rather than transformation efficiency [42]. Even though 0.5-1 cm long adventitious shoots were developed from some of the explants but these were not enough for DNA extraction.
Conclusion
In this study, it was concluded that the genetic transformation of Diamant, the commercial potato variety of Pakistan, which was carried out successfully by using the chitinase gene, may help in providing resistance against fungal phyto-pathogens and sustain its productivity. The transformed potato variety could also be used as fungus-resistant breeding material and offers new opportunities to develop improved potato cultivars for different agronomic and other desirable traits.
Acknowledgements
We generously thank Dr. Ikuo Nakamura, Chiba University, Japan for providing Streptomyces griseus strain HUT6037.
Conflict of Interest
There is no conflicts of interest.
References
- Paula A, Delamare L, Moschen-pistorello IT, et al. Antibacterial activity of the essential oils of Salvia officinalis L . and Salvia triloba L . cultivated in South Brazil. Food Chem 2007; 100: 603–608.
- Auld B. (2002). Book reviews. 518–521.
- Sharma N, Sharma KP, Gaur RK et al. Role of Chitinase in Plant Defense. Asian J Biochem 2011; 6(1): 29–37.
- Nagpure A, Choudhary B, Gupta RK. Chitinases: in agriculture and human healthcare. Crit Rev Biotechnol 2013; 34: 215-32.
- Devi P, Zhong H, Sticklen MB. In vitro morphogenesis of pearl millet [ Pennisetum glaucum (L.) R.Br.]: efficient production of multiple shoots and inflorescences from shoot apices. Plant Cell Rep 2000; 19: 546–550.
- Broglie K, Chet I, Holliday M, et al. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 1991; 254: 1194-1197.
- Jitonnom J, Lee VS, Nimmanpipug P, et al. Quantum mechanics/molecular mechanics modeling of substrate-assisted catalysis in family 18 chitinases: conformational changes and the role of Asp142 in catalysis in ChiB. Biochemi 2011; 50: 4697–4711.
- Bhattacharya D, Nagpure A, Gupta RK. Bacterial chitinases: properties and potential. Crit Rev Biotechnol 2007; 27: 21–28.
- Mendosa E.S, PHV. Aedes aegypti. Biotechnol Lett 1996; 4: 373–376.
- Hamid R, Khan MA, Ahmad M, et al. (2013). Chitinases: An update. J Pharm Bioallied Sci 2013; 5: 21–29.
- Mathivanan N, Kabilan V, Murugesan K. Purification, characterization, and antifungal activity of chitinase from Fusarium chlamydosporum, a mycoparasite to groundnut rust, Puccinia arachidis. Can J Microbiol 1998; 44: 646–651.
- Tsujibo H, Orikoshi H, Tanno H, et al. Cloning, sequence, and expression of a chitinase gene from a marine bacterium, Altermonas sp. strain O-7. J Bacteriol 1993; 175: 176–181.
- Jabeen N, Chaudhary Z, Gulfraz M, et al. Expression of Rice Chitinase Gene in Genetically Engineered Tomato Confers Enhanced Resistance to Fusarium Wilt and Early Blight. Plant Pathol J 2015; 31: 252–258.
- Das DK, Rahman A. Expression of a Rice Chitinase Gene Enhances Antifungal Response in Transgenic Litchi (cv. Bedana). American Journal of Plant Sciences 2018; 9: 2256-2275.
- Richa K, Tiwari IM, Devanna B, et al. Novel chitinase gene LOC_Os11g47510 from Indica Rice Tetep provides enhanced resistance against sheath blight pathogen rhizoctonia solani in rice. Front Plant Sci 2017; 8: 596.
- Marka R, Nanna RS. Optimization of factors affecting Agrobacterium-mediated genetic transformation in groundnut (Arachis hypogaea L.) Adv Plants Agric Res 2018; 8:275‒282
- Prasad K, Bhatnagar-Mathur P, Waliyar F, et al. Overexpression of a chitinase gene in transgenic peanut confers enhanced resistance to major soil borne and foliar fungal pathogens. J Plant Biochem Biotechnol 2013; 22: 222–233.
- Iqbal MM, Nazir F, Ali S, et al. Over Expression of Rice chitinase Gene in Transgenic Peanut (Arachis hypogaea L.) Improves Resistance Against Leaf Spot. Mol. Biotechnol. 2012; 50: 129–136.
- Kovács G, Sági L, Jacon G, et al. Expression of a rice chitinase gene in transgenic banana (’Gros Michel’, AAA genome group) confers resistance to black leaf streak disease. Transgenic Res 2013; 22(1): 117–130.
- Takahashi W, Fujimori M, Miura Y, et al. Increased resistance to crown rust disease in transgenic Italian ryegrass (Lolium multiflorum Lam.) expressing the rice chitinase gene. Plant Cell Rep 2005; 23: 811–818.
- Kishimoto K, Nishizawa Y, Tabei Y, et al. Detailed analysis of rice chitinase gene expression in transgenic cucumber plants showing different levels of disease resistance to gray mold (Botrytis cinerea). Plant Sci 2002; 162: 655–662.
- Ganesan M, Bhanumathi P, Kumari KG, et al. Transgenic Indian Cotton ( Gossypium hirsutum ) Harboring Rice Chitinase Gene ( Chi II ) Confers Resistance to Two Fungal Pathogens Am J Biochem Biotechnol 2009; 5: 63–74.
- Sreeramanan S, Maziah M, Xavier R. A protocol for Agrobacterium-mediated transformation of banana with a rice chitinase gene. Emirates J Food Agric 2009; 21: 18–33.
- Yamamoto T, Iketani H, Ieki H, et al. Transgenic grapevine plants expressing a rice chitinase with enhanced resistance to fungal pathogens. Plant Cell Rep 2000; 19: 639–646.
- Zhu H, Muthukrishnan S, Krishnaveni S, et al. Biolistic transformation of sorghum using a rice chitinase gene. J Genet Breed 1998; 52: 243–252.
- Li X, Sui X, Zhang Y, et al. An improved calcium chloride method preparation and transformation of competent cells. African J Biotechnol 2010; 9: 8549–8554.
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harb. Lab. Press. Cold Spring Harb. New York 2001.
- Zhang R, Hua LJ, Xiang CZ, et al. Cloning of a chitinase gene and the acquisition of transgenic potato plants. Acta Prataculturae Sinica 2009; 18: 51-58.
- Bin SG, Hong D, Hua LC, et al. (2009). Studies on Agrobacterium-mediated transformation of potato with ODREB2B gene. China Veget 2009; 16: 20-25
- Huang J, Ge X, Sun M. (2000). Modified CTAB protocol using a silica matrix for isolation of plant genomic DNA. Biotechniques 2000; 28: 432-434.
- Müller J, Krijgsman O, Tsoi J, et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat Commun 2014; 15(5): 5712.
- Shukla M, Jalil SU. Endochitinase: engineered resistance against fungal plant pathogens. Eur J Plant Pathol 2014; 105(4): 319-326.
- Ueda M, Kojima M, Yoshikawa T, et al. A novel type of family 19 chitinase from Aeromonas sp. No.10S-24. Cloning, sequence, expression, and the enzymatic properties. Eur J Biochem 2003; 270: 2513–2520.
- Hobel CFV, Hreggvidsson GO, Marteinsson VT, et al. Cloning, expression, and characterization of a highly thermostable family 18 chitinase from Rhodothermus marinus. Extremophiles 2005; 9: 53–64.
- Yano S, Rattanakit N, Wakayama M, et al. Cloning and expression of a Bacillus circulans KA-304 gene encoding chitinase I, which participates in protoplast formation of Schizophyllum commune. Biosci Biotechnol Biochem 2005; 69: 602–609.
- Yano S, Honda A, Rattanakit N, et al. Cloning and expression of chitinase A gene from Streptomyces cyaneus SP-27: the enzyme participates in protoplast formation of Schizophyllum commune. Biosci Biotechnol Biochem 2008; 72: 1853–1859.
- Punja ZK. Genetic engineering of plants to enhance resistance to fungal pathogens;a review of progress and future prospects. Can. J. Plant Pathol. 2001; 23: 216–235.
- Shin S, Mackintosh CA, Lewis J, et al. Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum. J Exp Bot 2008; 59: 2371–2378.
- Shah JM, Raghupathy V, Veluthambi K. Enhanced sheath blight resistance in transgenic rice expressing an endochitinase gene from Trichoderma virens. Biotechnol Lett 2009; 31: 239–244.
- Emani C, Garcia JM, Lopata-Finch E, et al. Enhanced fungal resistance in transgenic cotton expressing an endochitinase gene from Trichoderma virens. Plant Biotechnol J 2003; 1(5): 321–336.
- Bolar JP, Norelli JL, Wong KW, et al. Expression of Endochitinase from Trichoderma harzianum in Transgenic Apple Increases Resistance to Apple Scab and Reduces Vigor. Phytopathology 2000; 90: 72–77.
- Khan RS, Sjahril R, Nakamura I, et al. Production of transgenic potato exhibiting enhanced resistance to fungal infections and herbicide applications. Plant Biotechnol Rep 2008; 2: 13–20.

This work is licensed under a Creative Commons Attribution 4.0 International License.





