How to Cite | Publication History | PlumX Article Matrix
Raghdaa Shrief1, Reem Mohsen El-Kholy2, Mohamed Anies Rizk3 and Maysaa El Sayed Zaki3
1Medical Microbiology and Immunology Department, Mansoura Faculty of Medicine, Egypt.
2Department of Clinical Pathology, Menoufia Faculty of Medicine, Egypt.
3Clinical Pathology Department Mansoura Faculty of Medicine-Egypt.
Corresponding Author E-mail: may_s65@hotmail.com
DOI : http://dx.doi.org/10.13005/bbra/2739
ABSTRACT: The aim of the present study was to investigate the prevalence of tetracycline resistance genes among isolated S. aureus from healthcare associated surgical site infections. The present study included 350 clinical samples from healthcare associated surgical site infections. Identified S. aureus strains were subjected to antimicrobial susceptibility testing, detection of methicillin resistance by cefoxitin disc and molecular study of mecA and tet genes that were carried out by polymerase chain reaction and multiplex polymerase chain reaction, respectively. There were high resistance rates of isolated S. aureus to gentamicin (71.2%), kanamycin (66.5%) and ceftazidime (41.8%). Resistances to tetracycline, doxycycline and minocycline were 60.6%, 56.5% and 45.3%, respectively. In the comparison between MRSA and MSSA as regards antibiotics resistance, there was a significant increase in resistance to tetracycline, doxycycline, minocycline (P=0.0001) and erythromycin (P=0.01) among MRSA strains compared to MSSA. The tetracycline resistant genes detected were tetK (92.3%) and tetM (25.2%). Combined genes were detected in 22.3% of S. aureus. None of tetracycline isolates had tetL or tetO gene. There was significant higher frequency of telK, tetM and combined genes among MRSA compared to MSSA (P=0.0001). The present study highlights the prevalence of multiple antibiotics resistance among clinical isolates of S. aureus associated with healthcare associated infections. The resistance increases among methicillin resistant S. aureus. The resistance to tetracycline, minocycline and doxycycline were common. The common genetic basis of the resistance to tetracycline was the tetK and tetM genes.
KEYWORDS: mecA; S. Aureus; Tet genes
Download this article as:| Copy the following to cite this article: Shrief R, El-Kholy R. M, Rizk M. A, Zaki M. E. Prevalence of Tetracycline Resistant Genes in Staphylococcus aureus Isolates from Surgical Site Infections Egypt. Biosci Biotech Res Asia 2019;16(2). |
| Copy the following to cite this URL: Shrief R, El-Kholy R. M, Rizk M. A, Zaki M. E. Prevalence of Tetracycline Resistant Genes in Staphylococcus aureus Isolates from Surgical Site Infections Egypt. Biosci Biotech Res Asia 2019;16(2). Available from: https://bit.ly/2S9ZnEV |
Introduction
Staphylococcus is Gram positive bacterium that constitutes a major normal flora on the skin and mucous membrane. The virulent species is Staphylococcus aureus (S. aureus) that is associated with major hospital acquired infections such as surgical wound infections, sepsis and pneumonia (1).
Antibiotics therapy for S. aureus started with methicillin, which is formed of the phenol group of benzylpenicillin provided with two methoxy groups, that was active toward β-lactamase produced by S. aureus to penicillin. However, inappropriate use of antibiotics leads to emergence of methicillin-resistant S. aureus (MRSA) strains. The resistance is mainly related to the expression of penicillin-binding protein (PBP2a). The use of several classes of antibiotics resulted in the emergence of multi-resistant MRSA strains through the mutations in genes coding for target proteins and the acquisition and accumulation of antibiotic resistance genes (2). The gene associated with methicillin resistance is a mobile heterogenous gene, mecA, encoded by a Staphylococcal Cassette Chromosome mec (SCCmec) (3). SCCmec is formed of mecA gene with regulatory elements, a cassette chromosome recombinases (ccr) complex and the flanking direct repeat sequences that contain the integration site sequence for SCC. The classification of SCCmec is based upon the combination of ccr gene complex and the class of the mec gene (4-7).
Other antibiotics that are used for treatment of S. aureus especially in skin infections are tetracyclines. Nevertheless, there is an emergence of tetracycline resistance through the acquisition of tetracycline resistance (tet) genes and oxytetracycline resistance (otr) genes (8). Tetracycline resistance genes lead to resistance through active efflux pumps, ribosomal protection, and enzyme inactivation with 40 types of different resistant genes (9, 10).
Nowadays MRSA with resistant phenotypes to many antibiotics has emerged in healthcare associated infections. Understanding the molecular resistance mechanisms for these antibiotics is needed to develop active preventive measures for their spread.
The aim of the present study was to investigate the prevalence of tetracycline resistance genes among isolated S. aureus from healthcare associated surgical site infections at Mansoura University Hospitals.
Materials and Method
This study was a cross-sectional study that was carried out in Mansoura University hospitals from January 2017 till March 2018.
Specimens Collection
The wound swabs which were submitted to the microbiological laboratory from patients with healthcare associated infections according to CDC definition were included in the study (11). The study was approved by Mansoura Ethical committee and approvals were obtained from the participant patients.
Wound swabs were obtained from the patients by rolling the swabs over 1cm of the wound for 5 seconds then the swabs were transported in the sterile containers rapidly to the laboratory.
Specimens Processing
Swabs were cultured under aerobic conditions for 24 hours at 37°C. Colonies were identified by Gram stain then Gram positive cocci were identified by catalase, coagulase, and fermentation of mannitol tests (12). Identified S. aureus strains were subjected to antimicrobial susceptibility tests, detection of methicillin resistance by cefoxitin disc and molecular study of mecA and tet genes.
Antibiotics Susceptibility Testing by Discs Diffusion Method
The antibiotics susceptibility test was performed by discs diffusion method according to the Clinical Laboratory Standards Institute (CLSI) (13). The used discs were oxacillin (1 μg), gentamicin (10 μg), amikacin (30 μg), kanamycin (30 μg), erythromycin (15 μg), cefoxitin (5 μg), ceftazidime (30 μg), tetracycline (30 μg), doxycycline (30 μg), and minocycline (30 μg) (Oxoid-Thermal fisher). S. aureus ATCC 25923 was used as the control strain.
DNA Extraction
Pure colonies of S. aureus were prepared for DNA extraction by boiling method. Briefly, pure colonies were suspended in 100μl of sterile phosphate buffer and boiled at 100°C for 10 min in water bath then cooled. The suspension was kept frozen at -20°C till amplification procedures (14).
Polymerase Chain Reaction (PCR) for mecA Gene
The primers used for mecA amplification were listed in table 1. Five microns of the DNA extracted by the boiling method were added to total volume 100μl of amplification mixtures from Qiagen (Qiagen-Germany) with 1μM of mecA primers. The amplification conditions were as the following; an initial denaturation step at 94°C for 5 min followed by 40 cycles of amplification formed of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, and extension at 72°C for 60 sec ending with a final extension step at 72°C for 5 min. After PCR amplification, 5μl of amplicon were removed and subjected to agarose gel electrophoresis (1.5% agarose, 1× Tris-borate-EDTA buffer, 100 V, 40 min) to estimate the sizes of the amplification products by comparison with molecular marker 1kbp (15).
Multiplex PCR for Amplification of tetK, tetL, tetM Genes
Amplification of tet genes (tetK, tetL, tetM ) were performed using Qiagen PCR master kit (Qiagen). Multiplex PCR was prepared in a final volume of 25µl containing 0.5µM of each primer (Table 1). The PCR protocol consisted of an initial denaturation step at 94°C for 5 min then 30 amplification cycles at 94°C for 1 min, 51°C for 1 min and 72°C for 1 min, followed by a final extension step at 72°C for 5 min. Amplified products were analyzed by electrophoresis on 1.5% agarose gel containing 0.5 μg/ml ethidium bromide and photographed under UV illumination (16).
PCR for tetO
The primers used for amplification of tetO were listed in table 1. Amplification procedures were similar to the above except that the amplification cycles were 30 cycles at 94°C for 1 min, 57°C for 1min, 72°C for 1 min and a final extension for 72°C for 5 min. The amplified products were visualized as mentioned previously (16).
Table 1: Oligonucleotides sequences and sizes of different primers used in the amplification of mecA and tet genes.
| Size | Primers Sequences | Gene |
| 532 bp | F-AAAATCGATGGTAAAGGTTGGC | mecA |
| R-AGTTCTGCAGTACCGGATTTGC | ||
| 360 bp | F- GTAGCGACAATAGGTAATAGT | tetK |
| R- GTAGTGACAATAAACCTCCTA | ||
| 158 bp | F- AGTGGAGCGATTACAGAA | tetM |
| R- CATATGTCCTGGCGTGTCTA | ||
| 1077 bp | F-ATAAATTGTTTCGGGTCGGTAAT | tetL |
| R- AACCAGCCAACTAATGACAATGAT | ||
| 514 bp | F-AACTTAGGCATTCTGGCTCAC | tetO |
| R-TCCCACTGTTCCATATCGTCA |
Statistical Analysis
The statistical analysis was performed using SPSS 22.0 (SPSS Inc., Chicago, Illinois, USA). Descriptive data was expressed as number and percentage. Comparison was performed by Qi-square and P was considered significant if P was < 0.05.
Results
This study recovered 170 isolates of S. aureus from 350 wound swabs culture (48.6%) collected during the period of the study (15 months) (Figure 1).
Antibiotics sensitivity testing revealed high resistance rates of isolated S. aureus to gentamicin (71.2%), kanamycin (66.5%), Amikacin (52.3%) and ceftazidime (41.8%). Resistances to tetracycline, doxycycline and minocycline were 60.6%, 56.5% and 45.3%, respectively (Table 2).
Antibiotics resistance pattern of the isolates showed that 90 isolates (52.9%) were MRSA according to resistance to cefoxitin and/or oxacillin discs.
In the comparison between MRSA and methicillin sensitive S. aureus (MSSA) as regards antibiotics resistance, there was a significant increase in resistance to tetracycline, doxycycline, minocycline (P=0.0001) and erythromycin (P=0.01) among MRSA strains compared to MSSA (Table 3).
The tetracycline resistant genes detected were tetK (92.3%) (Figure 2) and tetM (25.2%). Combined tetK and tetM genes were detected in 22.3% of S. aureus resistant to tetracycline (Table 4). None of tetracycline resistant isolates had tetL or tetO gene. There was a significant higher frequency of either tetK, tetM or both genes among MRSA compared to MSSA (P=0.0001) (Table 5).
 |
Figure 1: Frequency of S. aureus strains isolated from healthcare associated surgical site infections.
|
Table 2: Antibiotics resistance pattern of the isolated S. aureus from surgical site infections.
| Antibiotics | S. aureus | |
| No. | % | |
| Gentamicin | 121 | 71.2% |
| Kanamycin | 113 | 66.5% |
| Tetracycline | 103 | 60.6% |
| Doxycycline | 96 | 56.5% |
| Cefoxitin | 90 | 52.9% |
| Oxacillin | 89 | 52.3% |
| Amikacin | 89 | 52.3% |
| Minocycline | 77 | 45.3% |
| Ceftazidime | 71 | 41.8% |
| Erythromycin | 66 | 38.8% |
| Total | 170 | 100% |
Table 3: Comparison of antibiotics resistance pattern between MRSA and MSSA strains.
| Antibiotics | MRSA (No.=90) | MSSA (No.=80) | P value | ||
| No. | % | No. | % | ||
| Amikacin | 50 | 55.6% | 39 | 48.7% | P=0.3 |
| Kanamycin | 56 | 62.2% | 57 | 71.3% | P=0.1 |
| Gentamicin | 65 | 72.2% | 56 | 70% | P=0.4 |
| Tetracycline | 90 | 100% | 13 | 16.3% | P=0.0001 |
| Minocycline | 70 | 77.8% | 7 | 8.7% | P=0.0001 |
| Doxycycline | 84 | 93.3% | 12 | 15% | P=0.0001 |
| Ceftazidime | 38 | 42.2% | 33 | 41.3% | P=0.5 |
| Erythromycin | 45 | 50% | 21 | 26.2% | P=0.01 |
Table 4: Frequency of tet genes among S. aureus strains resistant to tetracycline as detected by multiplex PCR.
| Gene | Frequency | |
| No. | % | |
| tetK | 95 | 92.3% |
| tetM | 26 | 25.2% |
| Combined tetK, tetM | 23 | 22.3% |
| Total | 103 | 100% |
Table 5: Distribution of tet genes among MRSA and MSSA strains as detected by multiplex PCR.
| Gene | MRSA (No.=90) | MSSA (No.=80) | P value | ||
| No. | % | No. | % | ||
| tetK | 81 | 90% | 14 | 17.5% | P=0.0001 |
| tetM | 22 | 24.4% | 4 | 5% | P=0.0001 |
| Combined tetK and tetM | 22 | 24.4% | 1 | 1.2% | P=0.0001 |
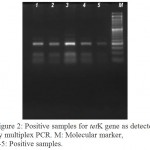 |
Figure 2: Positive samples for tetK gene as detected by multiplex PCR. M: Molecular marker, 1-5: Positive samples.
|
Discussion
Staphylococcus aureus represents a major pathogen that is related to healthcare associated infections. S. aureus, especially MRSA and multidrug resistant strains, is a common cause of surgical site infections (17).
In the present study, the etiology of about half of the isolates from healthcare acquired surgical site infections were due to S. aureus. Previous studies determined the high prevalence rate of S. aureus in surgical site infections up to 57% (17, 18). In several studies from Egypt, S. aureus was considered a common pathogen (61%) causing hospital acquired infections (71%) (19, 20).
MRSA represented 52.9% of the isolated S. aureus from surgical site infections in the present study. This represents a lower prevalence rate of MRSA to previous studies from Nepal, Egypt, Libya and Iran (17, 20-22). This may be attributed to the implantation of active surveillance and monitoring of patients and the rapid diagnostic tests, in addition to high orientation to preventive and control measures of MRSA during the period of the study in the hospital (23, 24). Moreover, the difference may be related to different clinical samples obtained from the patients, the duration of hospital stay and previous antimicrobial use.
MRSA strains in the present study were identified by phenotypic characters and molecular methods for detection of mecA gene. In agreement with our study, the molecular method for detection of mecA gene is an accurate and gold standard tool (25, 26). MRSA strains have resistance to methicillin and other semi-synthetic penicillinase resistant β-lactam antibiotics. The mecA gene is transferred horizontally and known to encode for a specific binding protein termed penicillin-binding protein 2a that has low affinity for β-lactam antibiotics which leads the complete synthesis of the bacterial cell wall without interruption by the β-lactam antibiotics (27, 28).
There were high resistance rates of isolated S. aureus to aminoglycosides and ceftazidime. Previous studies demonstrated multiple drug resistance among isolated S. aureus from healthcare associated infections (20). The high resistance to aminoglycosides among isolated S. aureus was similar to previous reports (20, 26, 29). The resistance pattern of isolated S. aureus depends upon the antibiotics policy in different institutions and infection control practices that lead to distribution diversity of antibiotics resistance genes.
In the present study, resistances to tetracycline, doxycycline and minocycline were 60.6%, 56.5% and 45.3%, respectively with significant increase in resistance to tetracycline, doxycycline, minocycline (P=0.0001) and erythromycin (P=0.01) in MRSA isolates compared to MSSA. These findings are in agreement with previous reports (16, 19). However, the resistance rate for tetracycline is higher than rates reported from the United States and Canada which have been reported to be 15.6% and 14.8%, respectively (30). Again, these findings reflect the difference in antibiotics policy between different geographic countries.
There are several mechanisms that lead to the development of tetracyclines resistance. One of these mechanisms is by the acquisition of tetK gene that leads to active efflux of divalent metal ion tetracycline from the bacterial cells (31). Another mechanism for tetracyclines resistance is mediated by tetM gene. This gene leads to tetracycline resistance through protection of the ribosome by the production of a ribosome protection protein that dislodges tetracycline from the ribosome freeing it from the inhibition by the drug (32).
In the present study, the tetracycline resistant genes detected were tetK (92.3%) and tetM (25.2%). Combined genes were detected in 22.3% of S. aureus, while none of the clinical isolates was positive for tetL and tetO genes. Similar previous studies have reported that tetK gene is the predominant gene of tetracycline resistance in clinical isolates of S. aureus followed by tetM (16, 33). However, other studies have documented that tetM is the predominant gene followed by tetK gene (8, 34). Similar to many reports, tetL and tetO were not detected in any of the isolates in the present study (16, 34).
Both tetK and tetM genes in the present work predominated in MRSA compared to MSSA. This is in accordance to previous findings (16, 33). On contrary, a literature has reported that tetM is more frequent in MRSA isolates (8).
These findings demonstrated that both tetM and tetK genes either single or combined have an important role in tetracycline resistance among MRSA strains in agreement with the previous report of Schmitz et al. (35). Similar to many reports, tetL and tetO were not detected in any of the isolates in the present study, indicating that these genes have no role in tetracycline resistance in our isolates (34, 36).
The predominance of tetM in tetracycline remittance MRSA may be attributed to previous exposure to tetracycline that leads to excision and transfer of the genetic element Tn916 carrying tetM gene (37). This theory may explain the relative less frequency of this gene among our isolates as the prescription of tetracycline is not common in our hospitals to S. aureus. On the other hand, tetK is transmitted through a small 4.4 kb plasmid known as pT181 from resistant strains in the absence of selective pressure (38).
Conclusion
The present study highlights the prevalence of multiple antibiotics resistance among clinical isolates of S. aureus associated with healthcare associated infections. The resistance increases among methicillin resistant S. aureus. The resistance to tetracycline, minocycline and doxycycline were common. The common genetic basis of the resistance to tetracyclines was the tetK and tetM genes.
Studying the mechanisms of antibiotics resistance is a mandatory tool for implementation of preventive and control measures to combat the resistant bacteria and reduce the risk of healthcare associated infections.
Conflicts of Interests
There is no conflicts of interests.
Funding Source
There is no funding source.
References
- DeLeo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest. 2009; 119(9):2464–74.
CrossRef - Stapleton PD, Taylor PW. Methicillin resistance in Staphylococcus aureus: mechanisms and modulation. Sci Prog. 2002; 85(1):57-72.
CrossRef - Ito T, Okuma K, Ma XX, Yuzawa H, Hiramatsu K. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist Updat. 2003; 6(1):41–52.
CrossRef - Hisata K, Kuwahara-Arai K, Yamanoto M, Ito T, Nakatomi Y, Cui L, Baba T, Terasawa M, Sotozono C, Kinoshita S, Yamashiro Y, Hiramatsu K. Dissemination of methicillin-resistant staphylococci among healthy Japanese children. J Clin Microbiol. 2005; 43(7):3364–72.
CrossRef - Kwon NH, Park KT, Moon JS, Jung WK, Kim SH, Kim JM, Hong SK, Koo HC, Joo YS, Park YH. Staphylococcal cassette chromosome mec (SCCmec) characterization and molecular analysis for methicillin-resistant Staphylococcus aureus and novel SCCmec subtype IVg isolated from bovine milk in Korea. J Antimicrob Chemother. 2005; 56(4):624–32.
CrossRef - Milheirico C, Oliveira DC, de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ’SCCmec IV multiplex’. J Antimicrob Chemother. 2007; 60(1):42–48.
CrossRef - Shore A, Rossney AS, Keane CT, Enright MC, Coleman DC. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob Agents Chemother. 2005; 49(5):2070–83.
CrossRef - Hui-Ling Ong M, Ho WY, Ng W-W, Chew C-H. High prevalence of tetM as compared to tetK among methicillin resistant Staphylococcus aureus (MRSA) isolates from hospitals in Perak, Malaysia. Jundishapur JMicrobiol.2017; 10(6) 1-6.
CrossRef - Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005; 245:195–203.
CrossRef - Hedayatianfard K, Akhlaghi M, Sharifiyazdi H. Detection of tetracycline resistance genes in bacteria isolated from fish farms using polymerase chain reaction. Vet Res Forum 2014; 5: 269-275.
- Tsai DM, Caterson EJ. Current preventive measures for health-care associated surgical site infections: a review. Patient Saf Surg. 2014; 8:42-55.
CrossRef - Reisner BS, Woods GL, Thomson RB Jr, Larone DH, Garcia LS, Shimizu RY. Specimen processing. In: Murray PR, Baron EJ, Pfaller MA, Tenover FC, Yolker RH (eds). Manual of Clinical Microbiology. 7th ed Washington DC, USA; 1999. pp. 64–104.
- Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing; M100-24: 2014. CLSI 24th Informational Supplement, Wayne PA, USA.
- Fatholahzadeh B, Hashemi FB, Emaneini M, Aligholi M, Nakhjavani FA, Kazemi B. Detection of Vancomycin Resistant Enterococci (VRE) isolated from urinary tract infections (UTI) in Tehran, Iran. Daru J Pharm Sci. 2006; 14:141–145.
- Meshref AA, Omer MK. Detection of (mecA) gene in methicillin resistant Staphylococcus aureus (MRSA) at Prince A/Rhman Sidery hospital , Al-Jouf, Saudi Arabia. J Med. Genet. Genomics 2011; 3 (3):41–45.
- Khoramrooz SS, Dolatabad SA, Dolatabad FM, Marashifard M, Mirzaii M, Dabiri H, Haddadi A, Rabani SM, Shirazi HRG, Darban-Sarokhalil D. Detection of tetracycline resistance genes, aminoglycoside modifying enzymes, and coagulase gene typing of clinical isolates of Staphylococcus aureus in the Southwest of Iran. Iran J Basic Med Sci. 2017; 20(8):912-919.
- Upreti N, Rayamajhee B, Sherchan SP, Choudhari MK, Banjara MR. Prevalence of methicillin resistant Staphylococcus aureus, multidrug resistant and extended spectrum β-lactamase producing gram negative bacilli causing wound infections at a tertiary care hospital of Nepal. Antimicrob Resist Infect Control, 2018; 7:121-131.
CrossRef - Bhowmik D, Chetri S, Paul D, Chanda DD, Bhattacharjee A. Detection and molecular typing of methicillin resistant Staphylococcus aureus from northeastern part of India. Med J Armed Forces India 2019; 75, 86 -89.
CrossRef - Ahmed EF, Gad GFM, Abdalla AM, Hasaneen AM, Abdelwahab SF. Prevalence of methicillin resistant Staphylococcus aureus among Egyptian patients after surgical interventions. Surg Infect. 2014; 15:404–411.
CrossRef - Abdel-Maksoud M, El-Shokry M, Ismail G, Hafez S, El-Kholy A, Attia E, Talaat M. Methicillin-Resistant Staphylococcus aureus recovered from healthcare- and community-associated infections in Egypt. Int J Bacteriol. 2016; 2016:5751-785.
CrossRef - Ghenghesh KS, Rahouma A, Tawil K, Zorgani A, Franka E. Antimicrobial resistance in Libya: 1970–2011. Libyan J Med. 2013; 27(8):1–8.
CrossRef - Jafari-Sales A, Farhadi F, Ezdiyadi M, Tarbiat-Nazloo D. Study of antibiotic resistance pattern in methicillin-resistant Staphylococcus aureus isolated from clinical samples of hospitals in Tabriz – Iran. Int J BioMed Public Health. 2018; 1(2):71-75.
- Chowers MY, Paitan Y, Gottesman B S, Gerber B, Ben-Nissan Y, Shitrit P. Hospital-wide methicillin-resistant Staphylococcus aureus control. Program: a 5-year follow-up. Infect Control Hosp Epidemiol. 2009; 30(8):778–781.
CrossRef - Hadler JL, Petit S, Mandour M, Cartter ML. Trends in invasive infection with methicillin-resistant Staphylococcus aureus, Connecticut, USA, 2001–2010. Emerg Infect Dis. 2012; 18(6):917–924.
CrossRef - Choi SM, Kim S-H, Kim H-J, Lee D-G, Choi J-H, Yoo J-H, Kang J-H, Shin W-S, Kang M-W. Multiplex PCR for the detection of genes encoding aminoglycoside modifying enzymes and methicillin resistance among Staphylococcus species. J Korean Med Sci. 2003; 18 (5):631–636.
CrossRef - Yadegar A, Sattari M, Mozafari NA, Goudarzi GR. Prevalence of the genes encoding aminoglycoside-modifying enzymes and methicillin resistance among clinical isolates of Staphylococcus aureus in Tehran, Iran. Microb Drug Resist. 2009; 15 (2):109–13.
CrossRef - Hartman BJ, Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984; 158 (2):513–6.
- Zapun A, Contreras-Martel C, Vernet T. Penicillin-binding proteins and beta-lactam resistance. FEMS Microbiol Rev. 2008; 32 (2):361–85.
CrossRef - Hauschild T, Sacha P, Wieczorek P, Zalewska M, Kaczyńska K, Tryniszewska E. Aminoglycosides resistance in clinical isolates of Staphylococcus aureus from a University Hospital in Bialystok, Poland. Folia Histochem. Cytobiol 2008; 46:225-228. 15:129-132.
CrossRef - Diekema D, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M and the SENTY Participants Group. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis. 2001; 32:S114-S132.
CrossRef - Jin J, Guffanti AA, Bechhofer DH, Krulwich TA. Tet(L) and tet(K) tetracycline-divalent metal/H+ antiporters: characterization of multiple catalytic modes and a mutagenesis approach to differences in their efflux substrate and coupling ion preferences. J Bacteriol. 2002; 184(17):4722–32.
CrossRef - Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001; 65(2):232–60.
CrossRef - Jones CH, Tuckman M, Howe AY, Orlowski M, Mullen S, Chan K, Bradford PA. resistance genes among Staphylococcus aureus isolates from phase 3 clinical trials of tigecycline for complicated skin and skin structure infections. Antimicrob Agents Chemother. 2006; 50:505-510.
CrossRef - Lim KT, Hanifah YA, Yusof M, Thong KL. ermA, ermC, tetM and tetK are essential for erythromycin and tetracycline resistance among methicillin-resistant Staphylococcus aureus strains isolated from a tertiary hospital in Malaysia. Indian J Med Microbiol. 2012; 30:203-207.
CrossRef - Schmitz FJ, Krey A, Sadurski R, Verhoef J, Milatovic D, Fluit AC; European SENTRY Participants. Resistance to tetracycline and distribution of tetracycline resistance genes in European Staphylococcus aureus isolates. J Antimicrob Chemother. 2001; 47 (2): 239-240.
CrossRef - Trzcinski K, Cooper BS, Hryniewicz W, Dowson CG. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2000; 45:763-770.
CrossRef - Su YA, He P, Clewell DB. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob Agents Chemother. 1992; 36(4):769–778.
CrossRef - Villedieu A, Diaz-Torres ML, Hunt N, McNab R, Spratt DA, Wilson M, Mullany P. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob Agents Chemother. 2003; 47(3):878–882.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





