How to Cite | Publication History | PlumX Article Matrix
Association of +10211T/G (Rs17846866) Variant of Adiponectin Gene With Type 2 Diabetes Mellitus
Mohammad Mustufa Khan1 and Roshan Alam2*
and Roshan Alam2*
1Department of Biomedical Sciences, School of Biological Engineering and Sciences, Shobhit University, Gangoh, Saharanpur, Uttar Pradesh, India- 247341
2Department of Biochemistry, Integral Institute of Medical Sciences and Research, Integral University, Lucknow, Uttar Pradesh, India- 226026
Corresponding Author E-mail: dr.roshan.alam@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2772
ABSTRACT: Various adiponectin gene (ADIPOQ) variants, located on chromosome 3q27 were associated with Type 2 diabetes mellitus (T2DM) in different ethnicity.In this study, it is aimed to find the association of +10211T/G (rs17846866) variant of ADIPOQ with T2DM and healthy controls in North Indians.In this study, 150 T2DM and 150 healthy control subjects aged between 25-75 years were recruited. Circulatory adiponectin levels were measured by commercially available ELISA kit. For genotype analysis, Polymerase chain reaction-restriction fragment length polymorphisms (PCR-RFLP) method was used.The genotypic analysis of rs17846866 variant of ADIPOQ has shown that there were no significant association of TT versus TG genotype (P=0.13) as well as TT versus GG genotype (P=0.11) with T2DM patients and healthy controls. However, the G allele frequency of the rs17846866 has shown significant association with T2DM (13.7%) as compared to healthy controls (7.7%, P=0.02). In T2DM, circulatory adiponectin level was significantly lower in TT genotype than TG genotypes (P=0.01). However, the circulatory adiponectin level was lower in GG genotype than TG genotype (P=0.49), but not significant.The result showed that rs17846866 variant of ADIPOQ was associated with altered circulatory adiponectin levels. The TT genotype may be the major contributor to reduce the circulatory adiponectin levels in T2DM. However, the G allele may be increased the risk of T2DM in North Indians.
KEYWORDS: ADIPOQ; Circulatory adiponectin; rs17846866; Type 2 diabetes; variant; Type 2 diabetes
Download this article as:| Copy the following to cite this article: Khan M. M, Alam R. Association of +10211T/G (Rs17846866) Variant of Adiponectin Gene With Type 2 Diabetes Mellitus. Biosci Biotech Res Asia 2019;16(3). |
| Copy the following to cite this URL: Khan M. M, Alam R. Association of +10211T/G (Rs17846866) Variant of Adiponectin Gene With Type 2 Diabetes Mellitus. Biosci Biotech Res Asia 2019;16(3).Available from: https://bit.ly/2BG7Zvk |
Introduction
Adiponectin was first identified in 1995, Scherer et al first characterized adiponectin as a novel 30-kDa secretory protein (Acrp30)1. Later in 1996, Nakano et al characterized adiponectin as mRNA transcripts of gelatin-binding protein of 28kDa size (GBP28)2 and Hu et al described adiponectin as a protein found highly expressed in adipose tissue (AdipoQ)3. Arita et al isolated adipose most abundant gene transcript 1 (apM1) and named it adiponectin4. Further studies demonstrated the multifactorial effects of adiponectin in adipocyte cells of adipose tissue and energy homeostasis which have a great impact on circulatory and storage sugar levels5, 6. Lara-Castro et al identified that it is a highly expressed transcript in preadipocytes differentiating into adipocytes7.
The circulating adiponectin levels in plasma have a wide range (2 to 30µg/ml), and that contributes about 0.01% of plasma proteins in adult individuals. The plasma level of this protein varies in different ethnicity. Indo-Asian has lower plasma adiponectin levels than Caucasians (3.3 to 4.9 µg/ml) and even than the Japanese population (5 to 10 µg/ml). Genome-Wide Association Studies (GWAS) among Asian and European populations identified ADIPOQ locus as the major gene for alteration in the circulatory adiponectin levels8, 9. ADIPOQ and its transcript has a versatile impact and linked multiple diseases such as diabetes10, metabolic syndrome11, obesity12, non-alcoholic fatty liver disease13, cardiovascular diseases14, neurodegenerative diseases15, and cancer16.
The ADIPOQ consists of 3 exons and 2 introns, contains about 16 kb of the genomic sequence, and located on chromosome 3q2717. We have included Intron 1 located +10211T/G (rs17846866) variant of ADIPOQ and associated with T2DM. Several studies reported that +10211T/G (rs17846866) variant of ADIPOQ contributed to T2DM18-20 (Figure 1).
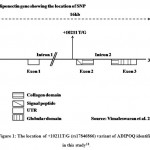 |
Figure 1: The location of +10211T/G (rs17846866) variant of ADIPOQ identified in this study18. |
Indians are highly susceptible to diabetes compared to other Europeans, Americans, and Asians21. It is an urgent need to find the possible genetic link of diabetes and its pathophysiology in a different region of India to reduce the burden of diabetes and its complication. Therefore, in this study, it is aimed to find the association between rs17846866 variant of ADIPOQ and T2DM.
Methods
Materials
Human blood samples
Subject selection
In this case-control study, 150 T2DM and 150 healthy controls were recruited aged between 25-75 years from outpatient diabetes clinic of Medical University. The study was carried out from May 2014 to October 2017. The study was approved by the ethical standards of the institutional committee. This study adhered to the principles of the 1964 Helsinki declaration and its later amendments or comparable ethical standards22. Written informed consent was taken from each subject recruited in the study.
Inclusion for T2DM subjects was done as per World Health Organization criteria (WHO)23, 24, whereas for healthy controls subjects who were unrelated and having no T2DM were included. Subjects with cardiovascular diseases, chronic liver disease, chronic kidney disease, any other infectious disease such as tuberculosis, chronic obstructive pulmonary disease, thyroid disorder, pregnant women were excluded for both (T2DM and healthy control) groups. Detailed medical history was taken from each subject.
The formula used for the sample size calculation
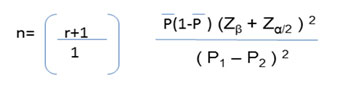
Where n = required sample size.; r = ratio of controls to cases.
P = average proportion exposed; Zβ = the desired power (0.84 for 80%); Zα = level of statistical significance (1.96); P1 = proportion of cases exposed (P1=0.35); P2 = proportion of control exposed (P2=0.20); n= 139≈150 per arm; Total sample size ≈300. Power of study is 80%.
Laboratory investigations
Total 5ml venous blood was withdrawn from each subject for genotyping analysis and biochemical investigation like FBS, PPBS, Lipid profile and Serum creatinine using commercially available ortho-clinical diagnostics kits (Johnson & Johnson) using Vitros-250 system chemistry autoanalyzer. HbA1c was measured by using whole blood EDTA samples on Bio-Rad D10 high-performance liquid chromatography (HPLC) analyzer (Bio-Rad, Hemel Hempstead, UK)25. Circulatory adiponectin levels were evaluated in serum using commercially available ELISA kit (USCN, Life Science Inc. Wuhan).
DNA extraction and Genotyping
DNA was extracted from human whole blood EDTA samples by using HiPurATM SPP Blood DNA Isolation Kit (Cat. No. MB541, HIMEDIA, India). Spectrophotometric analysis (Systronics-2205) and agarose gel electrophoresis using the Gel Doc system (Bio-Rad, Gel Doc XR+, Universal Hood II) revealed the concentration and the purity of the genomic DNA26. PCR-RFLP method was used for genotyping27. Primers were checked by using InSilico PCR online software. The rs17846866 (+10211T>G) variant of ADIPOQ was amplified using the forward primer, 5’-GCTAAGTATTACAGATTTCAGGGCAG-3’and the reverse primer, 5’-CAGCAACAGCATCCTGAGC-3’18. The PCR product size was detected 228bp. After that, the PCR products were digested with 10U of HinfI enzyme (New England Biolabs, UK), at 37°C for overnight. The restriction enzyme digested fragments of PCR products were observed on a 2.5% agarose gel. The wild type TT homozygote was detected as 101bp and 89bp fragments show HinfI enzyme restriction site absent. The mutant GG homozygote was detected as 134bp and 89bp fragments show HinfI enzyme restriction site present. The TG heterozygous contained the three fragments of 134bp, 101bp and 89bp (Figure 2).
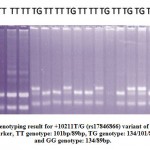 |
Figure 2: Genotyping result for +10211T/G (rs17846866) variant of ADIPOQ. M: Marker, TT genotype: 101bp/89bp, TG genotype: 134/101/89bp, and GG genotype: 134/89bp. |
Statistical Analysis
The IBM SPSS software version 20.0 (Armonk, NY, USA) was used to analyze the data. All the data were compared between the two groups by using the unpaired t-test. Values were given as a percentage (%) and mean ± SD (Standard Deviation). Odds ratio (OR) and 95% confidence interval (CI) were used to present allelic and genotypic frequencies and that were analyzed using χ2 test/Fischer’s exact test. For all data, P-value <0.05 was considered as statistically significant.
Results
In this study, 150 T2DM subjects with a mean age of (48.31±10.88 years) and disease duration (4.79±3.77 years), and 150 healthy control subjects with a mean age of (48.03±11.83 years) were recruited. For the anthropometric and biochemical profile of T2DM and healthy control subjects referred to Khan et al27.
The genotype and allele frequencies of the rs17846866 first intronic region variant of ADIPOQ in T2DM and healthy controls are shown in (Table 1). The frequencies of the TT, TG and GG genotypes of rs17846866 were 76%, 20.7%, 3.3% in T2DM and 85.3%, 14%, 0.7% in healthy controls, respectively. The allele frequencies of the T and G were 86.3%, 13.7% in T2DM and 92.3%, 7.7% in healthy controls, respectively. There were no significant association of homozygous TT and heterozygous TG genotype (OR: 0.60; CI: 0.33-1.11; P=0.13) as well as homozygous TT and GG genotype with T2DM and healthy controls (OR: 0.18; CI: 0.02-1.54; P=0.11). Although, T and G allele frequencies of the rs17846866 had a significant association with T2DM as compared to healthy controls (OR: 0.52; CI: 0.31-0.89; P=0.02).
Table 1: The genotypes and allele distribution of rs17846866 variant of ADIPOQ in T2DM and healthy controls
| rs17846866 Variant | T2DM
N (%) |
Healthy Controls
N (%) |
OR (95% CI) | P-value |
| Co-dominant
TT TG GG |
114 (76%) 31 (20.7%) 5 (3.3%) |
128(85.3%) 21 (14%) 1 (0.7%) |
1.00 0.60 (0.33-1.11) 0.18 (0.02-1.54) |
0.13 0.11 |
| Dominant
TT TG+GG |
114 (76%) 36 (24%) |
128(85.3%) 22 (14.7%) |
1.00 0.54 (0.30-0.98) |
0.06 |
| Recessive
TG+TT GG |
145 (96.7%) 5 (3.3%) |
149 (99.3%) 1 (0.7%) |
1.00 0.19 (0.02-1.69) |
0.21 |
| Alleles
T G |
259 (86.3%) 41(13.7%) |
277 (92.3%) 23 (7.7%) |
1.00 0.52 (0.31-0.89) |
0.02* |
*Significant considered as P<0.05.
Values are expressed as Number (N) and Percentage (%)
OR: Odd Ratio, CI: Confidence Interval
Clinical characteristics of the T2DM according to rs17846866 genotypes of ADIPOQ are shown in (table 2). There was a significant impact of rs17846866 genotype on circulatory adiponectin level, TG, and VLDL (P=0.02, 0.04, and 0.04, respectively). FBS, PPBS, HbA1c, and SCr were not significantly associated with rs17846866 genotype. However, PPBS and HbA1c were found significantly higher in TT genotype as compared to GG genotype (P=0.02, P=0.009, respectively). Parameters of obesity and metabolic syndrome i.e. WC, WHR, BMI, HDL, and LDL did not show any significant association with rs17846866 genotype. Circulatory adiponectin level was significantly lower in TT genotype than TG genotype (P=0.01). However, the circulatory adiponectin level was lower in GG genotype than TG genotype (P=0.49), but not significant.
Table 2: Clinical characteristics of T2DM with reference to rs17846866 genotypes of ADIPOQ
|
Parameters |
Genotypes | P-value | P-value | ||||
| TT (n=114) | TG (n=31) | GG (n=05) | TT vs. TG | TT vs. GG | TG vs. GG | ||
| FBS (mg/dl) | 163.89±49.83 | 153.32±44.14 | 129.80±36.08 | 0.29 | 0.13 | 0.27 | 0.26 |
| PPBS (mg/dl) | 253.13±77.85 | 233.68±75.61 | 169.60±36.74 | 0.22 | 0.02* | 0.07 | 0.21 |
| HbA1c (%) | 8.14±2.04 | 7.90±2.16 | 5.68±0.78 | 0.57 | 0.009* | 0.03* | 0.58 |
| TC (mg/dl) | 165.12±43.64 | 177.70±51.73 | 138.26±49.59 | 0.17 | 0.18 | 0.12 | 0.22 |
| TG (mg/dl) | 163.52±57.80 | 188.62±59.45 | 136.24±39.88 | 0.03* | 0.29 | 0.07 | 0.04* |
| HDL (mg/dl) | 39.00±11.36 | 39.11±11.47 | 40.72±20.58 | 0.96 | 0.75 | 0.79 | 0.96 |
| LDL (mg/dl) | 95.43±33.34 | 98.57±39.58 | 77.22±30.38 | 0.66 | 0.23 | 0.26 | 0.69 |
| VLDL (mg/dl) | 32.75±13.03 | 38.24±13.13 | 25.02±10.44 | 0.04* | 0.19 | 0.04* | 0.04* |
| SCr (mg/dl) | 2.17±1.35 | 2.49±1.42 | 3.28±1.51 | 0.25 | 0.08 | 0.26 | 0.27 |
| APN (μg/ml) | 1.78±0.85 | 2.25±1.03 | 1.90±1.08 | 0.01* | 0.76 | 0.49 | 0.02* |
| SBP (mmHg) | 139.66±25.97 | 141.65±27.01 | 160.00±31.77 | 0.71 | 0.09 | 0.18 | 0.72 |
| DBP (mmHg) | 82.87±15.63 | 80.61±14.60 | 82.40±21.14 | 0.47 | 0.95 | 0.81 | 0.45 |
| WC (cm) | 97.94±6.81 | 97.45±5.56 | 100.60±4.84 | 0.71 | 0.39 | 0.24 | 0.68 |
| WHR | 0.98±0.07 | 0.97±0.06 | 1.01±0.05 | 0.47 | 0.35 | 0.17 | 0.43 |
| BMI (kg/m2) | 24.94±4.83 | 25.10±4.15 | 24.42±4.15 | 0.87 | 0.81 | 0.74 | 0.86 |
*Significant considered as P<0.05.
Values are expressed as Mean ± Standard Deviation
FBS: Fasting Blood Sugar, PPBS: Post-Prandial Blood Sugar, HbA1c: Glycated Haemoglobin, TC: Total Cholesterol, TG: Triglyceride, HDL: High-Density Lipoprotein, LDL: Low-Density Lipoprotein, VLDL: Very Low-Density Lipoprotein, SCr: Serum Creatinine, APN: Adiponectin, SBP: Systolic Blood Pressure, DBP: Diastolic Blood Pressure, WC: Waist Circumference, WHR: Waist to Hip Ratio, BMI: Body Mass Index
Discussion
This study was performed to explain the role of genetic susceptibility to T2DM. Pathogenesis of T2DM is not much known. It is a multifactorial disease which develops by the interaction of genetic and environmental factors. Genetic factor includes multiple genes involved in development of T2DM. In the present study, we included rs17846866 variant of ADIPOQ to assess the genetic risk factor for T2DM. ADIPOQ variants were studied in different diseases28-30 as well as in T2DM31-33. We found a significant association of hypoadiponectinemia with T2DM. Similar results were reported in the literature in different ethnic group27, 33-34. Though, genetic studies were performed, limited and with inconsistent results. Alteration in circulatory levels of adiponectin was significantly associated with rs17846866 variant of ADIPOQ in T2DM. The rs17846866 variant dominant mode of inheritance showed nearly significant (TT vs. TG+GG, P=0.06) association with T2DM. Vimaleswaran et al. explained that the dominant mode of inheritance because the G allele (TG genotype) found a significantly higher risk than TT genotype while GG genotype did not have a higher risk than TG genotype. The G allele was found a significant association with T2DM in our results (P=0.02) and T allele is more common in healthy controls. Vimaleswaran et al found that TG genotype of rs17846866 had significantly higher risk for diabetes as compared to TT genotype and also associated with hypoadiponectinemia18. However, GG genotype has not associated with diabetes (OR: 0.18, CI: 0.02–1.54, P=0.11). The lack of GG genotype association with diabetes may be due to low frequencies of that GG genotype in both the T2DM and NHC groups. Saxena et al reported that rs17846866 variant of ADIPOQ increases the risk of T2DM19. Haplotype analysis and linkage disequilibrium study should be conducted between rs17846866 and other variants of ADIPOQ, because GG genotype and G allele of other variants of this gene reported a significant association with hypoadiponectinemia and T2DM35, 36. We observed that genotypic variation of rs17846866 variant of ADIPOQ had a significant association with hypoadiponectinemia and GG genotype have shown 1.5 folds higher serum creatinine as compared to TT genotype. This indicated that the ADIPOQ variant may be contributing the risk for diabetic nephropathy in T2DM patients. The previous study was reported that ADIPOQ variants have a strong correlation with the progression of diabetic nephropathy in T2DM patients37. Nazir et al have also found a significant positive association of ADIPOQ with diabetic nephropathy38. In addition, we observed that there was a significant impact of rs17846866 variant on TG, VLDL, and circulatory adiponectin levels. Raised TG and VLDL indicated dyslipidemia in T2DM patients.Hypoadiponectinemia and dyslipidemia in T2DM patients may contribute to increasing the risk for CVD. Verges reviewed that abnormalities of lipoprotein metabolism and increased TG and VLDL are one of the major risk factors for CVD in T2DM patients39. However, several studies have shown a discrepancy in the association of hypoadiponectinemia and CVD. Elevated circulatory adiponectin levels were independently linked with lower 10-year CVD risk in adults40. Similarly, Cheung et al were reported that ADIPOQ gene variant and hypoadiponectinemia is the independent predictor of CHD41. However, Menzaghi et al reviewed that high serum adiponectin is simply a marker of insulin sensitivity and glucose homeostasis, not a marker of CVD risk42. Similarly, the ADIPOQ variant associated with adiponectin levels was not associated with cardiometabolic risk factors43.
We did not find any association of WC, WHR, BMI, HDL, and LDL; this indicating that the variant of ADIPOQ included in our study is not associated with obesity or metabolic syndrome. The majority of study supported that ADIPOQ variants are associated with body weight, BMI and WHR i.e. obesity44, 45. Reasons for this is that the variant of ADIPOQ included in our study not associated with obesity but other variants of ADIPOQ gene is responsible for this30, 46.
Limitations and recommendations
Our study sample size might be smaller and we have taken only one variant rs17846866 of ADIPOQ. This study should be duplicated in a larger sample size with including other variants of ADIPOQ. Haplotype analysis and linkage disequilibrium study should be conducted between rs17846866 and other variants of ADIPOQ.
Conclusion
The result showed that rs17846866 variant of ADIPOQ was associated with altered circulatory adiponectin levels. The TT genotype may be the major contributor to reduce the circulatory adiponectin levels in T2DM. However, the G allele may be increased the risk of T2DM in North Indians.
Disclosure of Potential Conflicts of Interest
Funding
This study was not funded by any funding agency or company.
Conflict of Interest
Author M. M. Khan and R. Alam declare that they have no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed Consent
Written informed consent was obtained from all individual participants included in the study.
Acknowledgment
We are grateful to the residents of Medicine department and researchers of Biochemistry department including the technical staffs for their help and support in carrying out my thesis work. We also acknowledge Professor S. W. Akhtar, Founder, Integral University, Lucknow (India), for the invaluable help and financial support to carry out research work without any hindrance.
References
- Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes. J Biol Chem. 1995;270:26746–26749.
- Nakano Y, Tobe T, Choi-Miura NH, Mazda T, Tomita M. Isolation and characterization of GBP28, a novel gelatin-binding protein purified from human plasma. J Biochem. 1996;120:803–812.
- Hu E, Liang P, Spiegelman BM. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J Biol Chem. 1996;271:10697–10703.
- Arita Y, Kihara S, Ouchi N, Takahashi M, Maeda K, Miyagawa J et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun. 1999;257:79–83.
- Tao C, Sifuentes A, Holland WL. Regulation of glucose and lipid homeostasis by adiponectin: effects on hepatocytes, pancreatic β cells and adipocytes. Best Pract Res Clin Endocrinol Metab. 2014;28(1):43-58.
- Suyama S, Mekawa F, Maejima Y, Kubota Y, Kadowaki T, Yada T. Glucose level determines excitatory or inhibitory effects of adiponectin on arcuate POMC neuron activity and feeding. Sci Rep. 2016;6:30796.
- Lara-Castro C, Fu Y, Chung BH, Garvey WT. Adiponectin and the metabolic syndrome: mechanisms mediating risk for metabolic and cardiovascular disease. Curr Opin Lipidol. 2007;18:263–270.
- Heid IM, Henneman P, Hicks A, Coassin S, Winkler T, Aulchenko YS et al. Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: results of genome-wide association analyses including 4659 European individuals. Atherosclerosis. 2010;208(2):412-420.
- Ling H, Waterworth DM, Stirnadel HA, Pollin TI, Barter PJ, Kesaniemi YA et al. Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS Study. Obesity. 2009;17:737–744.
- Hsiao TJ, Lin E. A Validation Study of Adiponectin rs266729 Gene Variant with Type 2 Diabetes, Obesity, and Metabolic Phenotypes in a Taiwanese Population. Biochem Genet 2016;254(6):830-841.
- Kawada T. Serum adiponectin, its gene polymorphism and metabolic syndrome in adolescents. European Journal of Clinical Nutrition. 2016;70:645.
- Chang CS, Lu YJ, Chang HH, Hsu SH, Kuo PH, Shieh CC et al. Role of adiponectin gene variants, adipokines and hydrometry-based percent body fat in metabolically healthy and abnormal obesity. Obes Res Clin Pract. 2016;Pii: S1871-403X(16)30031-X.
- Du SX, Lu LL, Liu Y, Dong QJ, Xuan SY, Xin YN. Association of Adiponectin Gene Polymorphisms with the Risk of Coronary Artery Disease in Patients with Nonalcoholic Fatty Liver Disease in a Chinese Han Population. HepatMon. 2016;16(7):e37388.
- Chirumbolo S. Single Nucleotide Polymorphism (SNP) in the Adiponectin Gene and Cardiovascular Disease. Iranian Biomedical Journal. 2016;20(4):187-188.
- Yang Y, Hu W, Jiang S, Wang B, Li Y, Fan C et al. The emerging role of adiponectin in cerebrovascular and neurodegenerative diseases. Biochimica et Biophysica Acta. 2015;1852:1887–1894.
- Ye L, Wang G, Tang Y, Bai J. A population-specific correlation between ADIPOQ rs2241766 and rs1501299 and colorectal cancer risk: a meta-analysis for debate. Int J Clin Oncol. 2017;22(2):307-315.
- Kadowaki T, Yamauchi T. Adiponectin and Adiponectin Receptors. Endocrine Reviews. 2005;26(3): 439 – 451.
- Vimaleswaran KS, Radha V, Ramya K, Babu HNS, Savitha N, Roopa V et al. A novel association of a polymorphism in the first intron of adiponectin gene with type 2 diabetes, obesity and hypoadiponectinemia in Asian Human Genetics. 2008;123:599–605.
- Saxena M, Srivastava N, Banerjee M. Genetic association of adiponectin gene polymorphisms (+45T/G and +10211T/G) with type 2 diabetes in North Diabetes & Metabolic Syndrome: Clinical Research & Reviews. 2012;6:65–69.
- Ramya K, Ayyappa KA, Ghosh S, Mohan V, Radha V. Genetic Association of ADIPOQ gene variants with type 2 diabetes, obesity and serum adiponectin levels in south Indian population. Gene. 2013;532(2):253-62.
- NCDRFC: Non-Communicable Diseases Risk Factor Collaboration. Worldwide trends in diabetes since 1980: a pooled analysis of 751 population-based studies with 4·4 million participants. Lancet. 2016;387:1513–30.
- WMA: World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-4.
- WHO: World Health Organization. Definition and diagnosis of diabetes mellitus and intermediate hyperglycemia. Geneva, Switzerland: World Health Organization 2006. [http://apps.who.int/iris/bitstream/10665/43588/1/9241594934_eng.pdf]
- Khan MM, Sonkar GK, Alam R, Mehrotra S, Khan MS, Sonkar SK et al. Effect of age and body mass index on various clinical and anthropometric parameters of type 2 diabetic patients: a case-control study. Int J health Sci Res. 2016;6(11):132-142.
- Grant DA, Dunseath GJ, Churm R, Luzio SD. Comparison of a point-care analyser for the determination of HbA1c with HPLC method. Practical Laboratory Medicine. 2017;8:26-29.
- Ghatak S, Muthukumaran RB, Nachimuthu SK. A simple method of genomic DNA extraction from human samples for PCR-RFLP analysis. Journal of Biomolecular Techniques. 2013;24:224-231.
- Khan MM, Sonkar GK, Alam R, Singh S, Mehrotra S, Sonkar SK. Association of ADIPOQ Gene Variant rs266729 with Circulatory Adiponectin Levels in Patients with Type 2 Diabetes in North Indian Population: A Case-Control Study. Biomedical & Pharmacology Journal. 2017;10(1):407-417.
- Cheong MY, Bang OS, Cha MH, Park YK, Kim SH, Kim YJ. Association of the Adiponectin Gene Variations with Risk of Ischemic Stroke in a Korean Population. Yonsei Med J. 2011;52(1):20-25.
- Yu Z, Li W, Hou D, Zhou L, Deng Y, Tian M et al. Relationship between Adiponectin Gene Polymorphisms and Late-Onset Alzheimer’s Disease. PLoS ONE. 2015;10(4):e0125186.
- Riestra P, Gebreab SY, Xu R, Khan RJ, Bidulescu A, Correa A et al. Gender-specific associations between ADIPOQ gene polymorphisms and adiponectin levels and obesity in the Jackson Heart Study cohort. BMC Medical Genetics. 2015;16:65.
- Peters KE, Beilby J, Cadby G, Warrington NM, Bruce DG, Davis WA et al. A comprehensive investigation of variants in genes encoding adiponectin (ADIPOQ) and its receptors (ADIPOR1/R2), and their association with serum adiponectin, type 2 diabetes, insulin resistance and the metabolic syndrome. BMC Medical Genetics. 2013;14:15.
- Wang WL, Zhu H, Xie Y, Jin L. Relation between ADIPOQ gene polymorphisms and type 2 diabetes in a Chinese population. Int J Clin Exp Med. 2015;8(4):6124-6128.
- Goto A, Noda M, Goto M, Yasuda K, Mizoue T, Yamaji T et al. Plasma adiponectin levels, ADIPOQ variants, and incidence of type 2 diabetes: a nested case-control study. Diabetes research and Clinical Practice. 2017;127:254-264.
- Hivert MF, Manning AK, McAteer JB, Florez JC, Dupuis J, Fox CS et al. Common Variants in the Adiponectin Gene (ADIPOQ) Associated With Plasma Adiponectin Levels, Type 2 Diabetes, and Diabetes-Related Quantitative Traits. The Framingham Offspring Study. Diabetes. 2008;57:3353–3359.
- Lai H, Lin N, Xing Z, Weng H, Zhang H. Association between the level of circulating adiponectin and prediabetes: A meta-analysis. J Diabetes Invest. 2015;6:416–429.
- Sun P, Liu L, Chen J, Chen Y, Shi L, Imam MU et al. The polymorphism of rs266729 in adiponectin gene and type 2 diabetes mellitus: A Meta-Analysis. Medicine. 2017;96:47(e8745).
- Chung HF, Long KZ, Hsu CC, Mamun AA, Chiu YF, Tu HP et al. Adiponectin gene (ADIPOQ) polymorphisms correlated with the progression of nephropathy in Taiwanese male patients with type 2 diabetes. Diabetes Research and Clinical Practice. 2014;105(2):261-70.
- Nazir N, Siddiqui K, Al-Qasim S, Al-Naqeb D. Meta-analysis of diabetic nephropathy associated genetic variants in inflammation and angiogenesis involved in different biochemical pathways. BMC Medical Genetics. 2014;15:103.
- Verges B. Pathophysiology of diabetic dyslipidemia: where are we? Diabetologia. 2015;58(5):886-899.
- Kyrou I, Tsantarlioti O, Panagiotakos DB, Tsigos C, Georgousopoulou E, Chrysohoou C et al. Adiponectin circulating levels and 10-year (2002-2012) cardiovascular disease incidence: the ATTICA Study. Endocrine. 2017;58(3):542-552.
- Cheung CYY, Hui EYL, Cheung BMY, Woo YC, Xu A, Fong CHY et al. Adiponectin gene variants and the risk of coronary heart disease: a 16-year longitudinal study. European Journal of Endocrinology. 2014;171(1):107-115.
- Menzaghi C, Trischitta V. The adiponectin paradox for all-cause and cardiovascular mortality. Diabetes. 2018;67(1):12-22.
- Foucan L, Maimaitiming S, Larifla L, Hedreville S, Deloumeaux J, Joannes MO et al. Adiponectin gene variants, adiponectin isoforms and cardiometabolic risk in type 2 diabetic patients. Journal of Diabetes Investigation. 2014;5(2):192-198.
- Scotece M, Conde J, López V, Lago F, Pino J, Gómez-Reino JJ et al. Adiponectin and leptin: new targets in inflammation. Basic Clin Pharmacol Toxicol. 2014;114:97-102.
- Fisman EZ, Tenenbaum A. Adiponectin: a manifold therapeutic target for metabolic syndrome, diabetes, and coronary disease? Cardiovasc Diabetol. 2014;13:103.
- Wolski H, Krasnik W, Bogacz A, Weiczorek JB, Drews K, Greber A et al. Analysis of -11391G>A and +45T>G polymorphisms of ADIPOQ gene in women with excessive weight gain during pregnancy. Ginekol Pol. 2015;86:352-356.

This work is licensed under a Creative Commons Attribution 4.0 International License.





