How to Cite | Publication History | PlumX Article Matrix
1Department of Molecular Biology & Biotechnology, University of Kalyani, Kalyani, Nadia, India, 741235
2Department of Molecular Biology & Biotechnology, University of Kalyani, Kalyani, Nadia, India, 741235
Corresponding Author’s E-mail: utpal@klyuniv.ac.in
DOI : http://dx.doi.org/10.13005/bbra/2768
ABSTRACT: Poly (C) binding proteins (PCBPs) are members of sequence specific RNA binding protein family with conserved KH domain. There are four identified isoforms such as Pcbp1 or α-CP1 (α-Complex proteins), Pcbp2 or α-CP2, Pcbp3 or α-CP3 and Pcbp4 or α-CP4. Among them Pcbp1 and Pcbp2 are the most studied and found to be associated with various cellular functions such as transcriptional regulations, translational regulations and mRNA stability. Although two proteins share extensive similarity, they differ in function and localization. Pcbp1 has role in tumorigenesis, and metastasis, which are key phenomena of cancer. Role of pcbp2 has been well documented in the biology of RNA virus, namely translation and replication. Here, we studied expression pattern of Pcbp1 and Pcbp2 in three different cancer cell lines namely HeLa, RD, and A375 originated from different tissues. The results indicate not only differential abundance of these two proteins in three cell lines, but also discordant expression of pcbp1 in mRNA and protein level in three cell lines. The study therefore suggests post-transcriptional regulation of pcbp1 expression in these cell lines.
KEYWORDS: A375 Cells; Hela Cells; Immunoblotting; Poly (C) Binding Proteins; Real-Time PCR; RD Cells
Download this article as:| Copy the following to cite this article: Ghosh G, Basu U. Expression Pattern of Major Poly C Binding Protein (PCBP) Isoforms in Cancer Cell Lines of Cervix, Melanoma and Muscle. Biosci Biotech Res Asia 2019;16(3). |
| Copy the following to cite this URL: Ghosh G, Basu U. Expression Pattern of Major Poly C Binding Protein (PCBP) Isoforms in Cancer Cell Lines of Cervix, Melanoma and Muscle. Biosci Biotech Res Asia 2019;16(3). Available from: https://www.biotech-asia.org/?p=34188 |
Introduction
PCBPs are RNA binding proteins with domains that bind to single stranded nucleic acids in sequence specific manner. PCBPs are transcribed at four dispersed loci and named pcbp1 or α-CP1, pcbp2 or α-CP2, pcbp3 or α-CP3 and pcbp4 or α-CP4 [1]. They share three evolutionary conserved KH domains, responsible for poly C binding. Two of them are located near N-terminus and one near C-terminus. Highest sequence conservation is found between these KH domains but sequence conservation is also evident in inter-KH domains [1, 2 ]. Gene duplication and retrotransposition are the two events that lead to formation of PCBP2, PCBP3, PCBP4 and PCBP1 proteins [1, 3]. Retrotransposition of PCBP2 splice product generated intronless PCBP1 loci [1, 3, 4]. PCBP1 and PCBP2 are the most studied isoform among the four PCBPs. Both of them are attributed to different cellular functions such as transcriptional regulations, translational regulations and mRNA stability [5, 6]. PCBP1 is negative regulator of CD44 splicing in HepG2 cells and hence loss of PCBP1 contributes to metastatic phenotype of human hepatic tumour [7]. Knocking down of PCBP1 enhances metastatic tumour formation, as PCBP1 is the translational repressor of metastatic PRL-3 protein [8]. Repression of PCBP1 enhances tumour formation by attenuating P27kip1mRNA stability and translation [9]. PCBP2 depletion induces apoptosis in gastric cancer cells [10]. PCBP2 knock down inhibits glioma growth in vitro and in vivo by inducing caspase3- mediated apoptosis [11]. Therefore, certainly PCBP1 and PCBP2 repression in cancer cells play important roles in tumorigenesis and metastasis.
Role of pcbp family members in post-transcriptional regulation of cellular mRNA has been documented. In muscle, PCBP1 interacts with argonaute 2 and modulates miRNA expression [12]. Translational silencing of 15-lipoxygenase mRNA in early erythroblasts is mediated through a complex containing pcbp1 formed at CU rich motif within its 3’UTR [13].
In poliovirus transcription-translation switch is based on pcbp. Binding of pcbp2 at 5´ of poliovirus RNA genome is essential for its translation [14]. Actually, poliovirus RNA tranalation is mediated through internal ribosome entry site (IRES), not through 5´ cap. PCB2 binding to type I IRES activates translation and this effect is not limited to poliovirus only. Type I IRESs and even structurally divergent cadicivirus IRES function in PCBP2 dependent manner [15].
However, regulation of Poly (C) Binding proteins expression is not well documented. In the present study we therefore, studied the expression pattern of PCBP1 & 2 in three different cell lines that are derived from three different human tissues. We chose HeLa derived from human cervical cancer cells, RD derived from human Rhabdomyosarcoma cells and A375 derived from human skin malignant melanoma cells. We isolated total RNA from these cells and cDNA prepared from these samples were analysed by Q-PCR to get mRNA expression pattern of PCBP1 and PCBP2. Then we cell lysate from these cells and immunoblotting was done to check protein level expression pattern of the same. Our results demonstrate that both Pcbp1 and Pcbp2 transcripts are more abundant in HeLa cells compared to RD and A375. Interestingly there is no significant difference in the protein level of PCBP1 among three cell lines, whereas PCBP2 is more abundant in HeLa, compared to RD and A375 cells and least abundantin A375 cells. Therefore, mRNA-protein mismatch for Pcbp1 in HeLa, is quite evident suggesting post-transcriptional regulation of its expression in these tested cell lines.
Materials and Methods
Cell Culture
Monolayered culture of HeLa, RD and A375 cells were maintained in DMEM (Himedia) containing streptomycin (100µg/ml), neomycin (50µg/ml), and supplemented with 10% FBS (Invitrogen).
RNA isolation and cDNA preparation
Total RNA from all three cells was isolated from 90% confluent 35mm plates using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Then the RNA samples were treated with DNaseI (NEB) at 37 ºC for 30 mins in a heat block. After that, RNA was extracted with phenol- chloroform (1:1) and precipitated with 1/10th volume of Na-acetate and 0.8th volume of isopropanol. RNA Pellet was washed with 75% ethanol and finally dissolved in nuclease free water. 1µg of total RNA from each sample was used for cDNA preparation. Frist strand cDNA synthesis was done by MMLV Reverse Transcriptase (Epicentre) using oligo dT (Invitrogen) as primer according to the manufacturer’s protocol. cDNA from all samples were stored at -20ºC.
Real-Time PCR
Real-Time PCR was done using iTaq™ Universal SYBR® Green Supermix (Bio-Rad) according to manufacturer’s protocol. Primer ( Table 1) concentration was 2 pmol and 0.5 µl of cDNA from each sample was used as template for every 20 µl reaction.
Table 1: Fold difference in the expression of pcbp1 and pcbp2 for all these cell lines were calculated with 2–∆∆CT method using β-actin as endogenous control [16].
|
Gene |
Forward Primer |
Revers Primer |
| β-Actin | 5’TGCCGACAGGATGCAGAAG 3’ | 5’GCCGATCCACACGGAGTACTT 3’
|
| Pcbp1 | 5’ACGGAAAGGAAGTAGGCAGCAT 3’ | 5’GCAGTTCCCCTCCGAGATGT 3’ |
| Pcbp2 | 5’CCGGAGCCCAAGGCTTTA 3’
|
5’AAACCTTGAAATATAACACTCCATGCA 3’
|
Cell Lysate Preparation and Immunoblotting
90% confluent 35mm plates were used for protein isolation. After aspirating media, cells were washed with ice cold 1X PBS and NP40 buffer (150 mm NaCl, 50mm Tris pH 8.0, 1% NP-40) supplemented with 1X Protease inhibitor cocktail (Complete mini, Roche) was added. Cells were then scrapped and kept on ice for 30 mins, centrifuged at 14000 rpm for 30 mins at 4 ºC. Supernatant was transferred to fresh microcentrifuge tube. Protein estimation was done by 1X Bradford reagent (Bio-Rad). 50 µg of protein was cooked with 1X SDS sample buffer at 70 ºC for 10 mins and resolved in 12% poly acrylamide gel with 1X (24 mM Tris, 250 mM glycine, 0.05% SDS) Tris-Glycine running buffer. Then protein was transferred onto PVDF membrane over night at 150 mA constant current at 4 ºC using ice cold transfer buffer (192 mM glycine, 25 mM Tris pH 8.3, 20% methanol, 0.05% sodium dodecyl sulphate). Membrane was blocked in 5% BSA-1X TBS (Tris buffered saline or TBS; 150 mM NaCl, 50 mM Tris pH 7.5) solution for 1hour at 4 ºC. Membrane was incubated with rabbit anti α-Tubulin (CST) and rabbit anti Pcbp2(Abcam) antibody mix (dilution 1:1000 each) at room temperature for an hour followed by 1X TBST (1X-TBS+ 0.05% Tween 20) washing five times changing buffer in every 5 mins. Membrane was then incubated with HRP-conjugated anti rabbit secondary antibody (milipore; dilution 1:5000) for 1 hour at room temperature followed by 1X TBST washing same as before. Membrane was incubated with ECL solution and chemiluminiscence was photographed by ChemiDoc XRS+ system (Bio-Rad). The same membrane was the stripped in mild stripping buffer (200 mM glycine, 0.1% SDS, 1% tween 20), blocked and then reprobed with rabbit anti pcbp1 (Abcam) antibody (dilution 1:1000) as before. ImageJ software (http://rsbweb.nih.gov/ij/index.html) was used for band density quantification.
Results and Discussion
Many reports suggest that PCBP plays crucial role in many physiological phenomena including carcinogenesis and metastasis. We therefore aimed to investigate the expression profile of pcbp1 and 2 in three malignant cell lines from different tissue types. HeLa is human ovarian cancer cell line, whereas RD is derived from sarcoma and A375 is originated from malignant melanoma. mRNA expression of pcbp1 and pcbp2 in cell lines HeLa, RD and A375 was studied through quantitative PCR. Figure1 shows real-time experimental data exhibiting their relative abundances in all three cell lines. pcbp1 and pcbp2 mRNA are most abundant in HeLa cells in comparison with RD and A375. There is no significant difference in transcript level of pcbp2 in RD and A375 cells but mRNA expression of Pcbp1 differs significantly in RD and A375 cells. Having found that the relative expression of pcbp1 and 2 transcripts vary between cell lines, we studied the expression of these two genes in the protein level. The lysate of cultured cells were analysed through SDS-PAGE followed by immunoblot using antibodies specific for PCBP1 and PCBP2. Although, pcbp1 transcript is found to be higher in HeLa cells compared to RD and A375, no significant difference in its protein level among the three chosen cell lines has been observed. PCBP2 western analysis shows that it is most abundant in HeLa cells and least abundant in A375 cells. Therefore, in contrast to pcbp2, there is lack of correlation between pcbp1 in mRNA and protein level in HeLa, RD and A375 cells. Based on this observation, we propose that there may be post-transcriptional regulation of pcbp1 expression in these cells lines.
Expression of protein of a gene can be controlled in many steps. Although transcription is the most common to regulate protein expression, post-transcriptional regulation of expression has been documented for many genes. Stability of transcript and rate of translation initiation are two major parameters that control expression of the protein coded by an mRNA. Stability of mRNA, as indicated by its half life can vary from minutes to days. Lower stability of the transcripts is mainly due to their cleavage by ribonucleases, whereas stable mRNAs are capable of resisting RNases. The resistance towards RNases come from their interaction with proteins that bind either sequence specific or structure specific manner. The rate of translation of an RNA can be controlled by interacting proteins, RNA secondary structural elements and miRNA targeting its 3´UTR [17]. Results of the present study since suggest post-transcriptional regulation of pcbp1, in the tested cells at least any of the above mentioned regulatory mechanism may play its role in the expression of pcbp1.
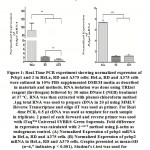 |
Figure 1: Real-Time PCR experiment showing normalized expression of Pcbp1 and 2 in HeLa, RD and A375 cells: HeLa, RD and A375 cells were cultured in 10% FBS supplemented DMEM media as described in materials and methods. |
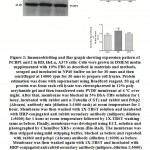 |
Figure 2:. Immunoblotting and Bar graph showing expression pattern of PCBP1 and 2 in RD, HeLa, A375 cells. |
 |
Figure 3:Schematic diagram showing regulation of poly C binding protein 1 in HeLa, RD and A375 cells. |
Conclusion
Quantitative PCR and immunoblotting experiment demonstrated differential mRNA and protein level expression of poly C binding protein 1 and 2 in HeLa, RD and A375 cells. We found that poly C binding protein 1 is most abundant in HeLa and least abundant in A375 cells in transcript level. Pcbp2 transcript is highest in HeLa cells but in other two cells, the transcript level does not significantly vary. Interestingly, PCBP1 protein is equally abundant in all three cell lines whereas PCBP2 protein level distribution is highest in HeLa and lowest in A375 cells. Unlike pcbp2, there is mismatch in expression of pcbp1 in mRNA and protein level. Therefore, our result suggests that pcbp1 expression in HeLa, RD and A375 may be post-transcriptionally regulated (Figure 3).
Acknowledgement
We acknowledge University of Kalyani DST-Purse programme for their financial support. University of Kalyani and Department of Molecular Biology and Biotechnology for the instrumentation and laboratory facilities.
Funding Source
DST-Purse Programme
Personal Research Grant to Utpal Basu and Gargi Ghosh
Reference
- Makeyev A. V and Liebhaber S. A. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 2002; 8:265–278
CrossRef - Makeyev A. V and Liebhaber S. A. Identification of Two Novel Mammalian Genes Establishes a Subfamily of KH-Domain RNA-Binding Proteins. Genomics 2000; 67(3): 301-316
CrossRef - Ghanem L. R, Kromer A, Silverman I. M, Chatterji P and Traxler E, Penzo-Mendez A, Weiss M. J, Stanger B. Z and Liebhaber S. A. The Poly(C) binding protein Pcbp2, and its retrotransposed derivative Pcbp1, are 2 independently essential to mouse development. Mol. Cell. Biol. 2016; 36(2): 304–319
CrossRef - Makeyev A. V, Chkheidze A. N and Liebhaber S. A. A set of highly conserved RNA-binding proteins, alphaCP-1 and alphaCP-2, implicated in mRNA stabilization, are coexpressed from an intronless gene and its intron-containing paralog. J. Biol Chem. 1999; 274(35):24849-57
CrossRef - Yeap B. B, Voon D. C, Vivian J. P, McCulloch R. K, Thomson A. M, Giles K. M, Czyzyk-Krzeska M. F, Furneaux H, Wilce M. CJ, Wilce J. A, and Leedmana P. J. Novel Binding of HuR and Poly(C)-binding Protein to a Conserved UC-rich Motif within the 3-Untranslated Region of the Androgen Receptor Messenger RNA. JBC 2002; 270(30): 27183–27192
CrossRef - Choi H. S, Hwang C. K, Song K. Y, Law P, Wei L, and Loh H. H. Poly(C)-binding Proteins as Transcriptional Regulators of Gene Expression. Biochem Biophys Res Commun. 2009; 380(3): 431–436
CrossRef - Zhang T, Huang XH, Dong L, Hu D, Ge C, Zhan YQ, Xu WX, Yu M, Li W, Wang X, Tang L, Li CY. Xiao-Ming YanPCBP-1 regulates alternative splicing of the CD44 gene and inhibits invasion in human hepatoma cell line HepG2 cells. Molecular Cancer 2010; 9:72
CrossRef - Wang H, Vardy L. A, Tan C. P, Loo J. M, Guo K, Li J, Lim S. G, Zhou J, Chng W. J, Ng S. K, Li H. X, and Zeng Q. PCBP1 Suppresses the Translation of Metastasis-Associated PRL-3 Phosphatase. Cancer Cell. 2010; 18(1):52-62
CrossRef - Shi H, Li H, Yuan R, Guan W, Zhang X, Zhang S, Zhang W, Tong F, Li L, Song Z, Wang C, Yang S and Wang H.
PCBP1 depletion promotes tumorigenesis through attenuation of p27Kip1 mRNA stability and translation. JECCR 2018; 37:187
CrossRef - Ji FJ, Wu1 YY, An Z, Liu XS, Jiang JN, Chen FF and Fang XD. Expression of both poly r(C) binding protein 1 (PCBP1) and miRNA-3978 is suppressed in peritoneal gastric cancer metastasis. Sci Rep 2017; 7(1):15488
CrossRef - Han W, Xin Z, Zhao Z, Bao W, Lin X, Yin B, Zhao J, Yuan J, Qiang B, Peng X. RNA-binding protein PCBP2 modulates glioma growth by regulating FHL3. J Clin Invest 2013; 123(5):2103-18
CrossRef - Espinoza-Lewis, R. A., Yang, Q., Liu, J., Huang, Z. P., Hu, X., Chen, D., & Wang, D. Z. (2017). Poly (C)-binding protein 1 (Pcbp1) regulates skeletal muscle differentiation by modulating microRNA processing in myoblasts. Journal of Biological Chemistry, 292(23), 9540-9550.
CrossRef - Ostareck, D. H., Ostareck-Lederer, A., Wilm, M., Thiele, B. J., Mann, M., & Hentze, M. W. (1997). mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell, 89(4), 597-606.
CrossRef - Parsley, T. B., Towner, J. S., Blyn, L. B., Ehrenfeld, E., & Semler, B. L. (1997). Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. Rna, 3(10), 1124-1134.
- Asnani, M., Pestova, T. V., & Hellen, C. U. (2016). Initiation on the divergent Type I cadicivirus IRES: factor requirements and interactions with the translation apparatus. Nucleic acids research, 44(7), 3390-3407.
CrossRef - Livak K. J and Schmittgen T. D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. METHODS. 2001; 25: 402–408
CrossRef - Suay, L., Salvador, M. L., Abesha, E., & Klein, U. (2005). Specific roles of 5′ RNA secondary structures in stabilizing transcripts in chloroplasts. Nucleic acids research, 33(15), 4754-4761.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.






