How to Cite | Publication History | PlumX Article Matrix
Isolation and Characterization of Biosurfactant Producing Bacteria from oil-contaminated Water
Ayman Youssef Ibrahim Ewida* and Walaa Salah El-din Mohamed
and Walaa Salah El-din Mohamed
Department of Microbiology, National Water Research Center, Egypt.
Corresponding author email: aymanyi@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2801
ABSTRACT: Biosurfactants are chemical compounds produced by some microorganisms to initiate oil biodegradation. They have been applied generously in many industries. The present work aimed to isolate and identify a new bacterial strain, of water habitat, capable of producing biosurfactant. So, water samples were collected from three different water environments including river Nile at Alkanater city, Qalyubia governorate; was representing clear raw water. River Nile at ship settlement station, Imbaba city, Giza governorate; was representing oil- contaminated water, where there were some oil spills from ship fixation. Rahawy drain, Giza governorate; was representing highly polluted wastewater. The bacterial community of each water environment was isolated and inventoried, then screened for biosurfactant production by blood hemolysis, oil spreading technique, drop collapse assay, foaming activity and emulsification activity. Bacterial strains isolated from the oil-contaminated environment showed high potential for biosurfactant production, and the best biosurfactant producing isolate was identified by 16S rRNA technique as Pseudomonas protegens, and the produced biosurfactant was belong to rhamnolipid group.
KEYWORDS:
Bacteria; Biosurfactant; Pseudomonas protegens; Rhamnolipid; Water
Download this article as:| Copy the following to cite this article: Ewida A. Y. I, Mohamed W. S. E. Isolation and Characterization of Biosurfactant Producing Bacteria from oil-contaminated Water. Biosci Biotech Res Asia 2019;16(4). |
| Copy the following to cite this URL: Ewida A. Y. I, Mohamed W. S. E. Isolation and Characterization of Biosurfactant Producing Bacteria from oil-contaminated Water. Biosci Biotech Res Asia 2019;16(4). Available from: https://bit.ly/38X0yzt |
Introduction
Surfactants are organic substances which decrease the tension between two liquids, or between a liquid and a solid. They are mainly containing two parts; a hydrophobic part which is less soluble in water, and having long-chain of fatty acid; a hydrophilic part which is more soluble and containing carbohydrate or carboxylic acid [1]. Biosurfactants are the same like chemical surfactants; they are surface active compound, but synthesized by microbes like fungi, yeast and bacteria [2].
Biosurfactants are characterized as non-toxic material and have specific properties of dropping surface tension, stabilizing emulsions, and promoting foaming [3]. They are used in several industries such as pharmaceuticals, cosmetics, detergents and food preservatives [3]. Recently, interest in biosurfactants production has increased due to their economic benefits, flexibility, low toxicity, relative ease of preparation and more eco-friendly than chemical surfactant.
Glycolipids are mostly biosurfactants, which are lipids bound to a carbohydrate. The relation is either an ether group or an ester bond. Rhamnolipids are among the best known glycolipids. Many species of Pseudomonas have produced rhamnolipids and have significant antimicrobial activity against several different microorganisms. They contain one or two β-hydroxydecanoic acid molecules bound to two rhamnose molecules. There are some other structures of glycolipid biosurfactants including trehalolipids which is produced by some gram-positive bacteria, and sophorolipids which is produced by some yeast [4-6].
Several bacterial genera such as Bacillus, Burkholderia, Flavobacterium and Pseudomonas are reported to produce biosurfactants, in general, microorganism are considered as generously biosurfactant producers [7]. Oil contaminated environments are rich in biosurfactant producing bacteria, where they generating biosurfactants to use hydrocarbons as a carbon source. That often mineralizes them or transforms them into harmless products [8].
A combination of various screening methods for detection of potential biosurfactant producers is required, such as; haemolytic activity, oil spreading assay, drop collapse assay, foaming activity and emulsification assay [9].
The objectives of the present study are: to isolate and screen biosurfactant producing bacteria from different water environments; to carry out a comparative study concerning the ability of a given bacterial isolate to produce biosurfactant with the water quality of its origin; to identify the bacterial strains showing their potential to produce biosurfactant; to identify the biosurfactant type.
Materials and Methods
Sampling sites and water quality assessment studies
Water samples were collected from different water environments (3 sites). The first site was the river Nile at Alkanater city, Qalyubia governorate (named site 1) representing clear raw water. The second site was river Nile at ship settlement station, Imbaba city, Giza governorate (named site 2) representing oil- contaminated water, where there were some oil spills from ship fixation. Finally, the third site was Rahawy drain, Giza governorate (named site 3) representing highly contaminated wastewater. All samples were collected and transported to the laboratory for analyses according to the Standard Methods for the Examination of Water and Wastewater [10].
Isolation of biosurfactant producing bacteria
Isolation of bacterial community of each water environment
Water samples were serially diluted from 10-1 to 10-6, 1 ml of 10-4 to 10-6 dilutions were used to inoculate plate count agar plates by spread plate method. All the plates were incubated at 35 oC for 24 h. According to the cultural differentiation, colonies were characterized, isolated, and purified. Each bacterial isolate was given a special code to be maintained for further investigations.
Screening of isolates for biosurfactant production
Blood hemolysis, oil spreading technique, drop collapse assay, foaming activity and emulsification activity were the screening tests used for detection of biosurfactant production potential for each bacterial isolate [9, 11, and 12].
Blood hemolysis
The hemolytic activity was the first screening test to identify biosurfactant producing bacteria [13]. Blood agar plates were prepared by adding 5 ml sheep blood on 1 L nutrient agar medium. Freshly prepared colonies were streaked on blood agar plates and incubated for 72 h. at 30 °C. The presence of greenish color or clear zone around the colonies (α- or β- hemolysis) indicating that the bacterial isolate under investigation may has biosurfactant production ability [12, 14].
Oil Spreading Technique
Bacterial isolates were inoculated, separately, into sets of 10 ml of broth media and incubated at 30 °C for 72 h. Supernatants were collected by centrifuging culture media at 3000 rpm for 30 min. They will be used for the various biosurfactant screening tests. The oil spreading test was carried out by adding 1 ml of vegetable oil on the surface of 30 ml of distilled water (contained into a petri dish bottom). On the center of the oil layer, 10 μl of the culture supernatant were gently added, and the observations were recorded after 1 min. The presence of biosurfactant will cause oil displacement, and a clear zone will appear [15]. The displacement diameter was measured in (mm), known as oil-displacement activity. Replicates for each isolate were carried out, and a water drop was used as a negative control.
Drop Collapse Assay
Carefully, a drop of the culture supernatant was placed on an oil coated glass slide, and observations were recorded after 1 min. If the drop of the culture supernatant collapsed and spread on the oil coated surface, biosurfactant was considered present. Negative control was also carried out using distilled water [16].
Foaming activity
Each bacterial isolate was used to inoculate 100 ml of nutrient broth medium and incubated at 30 °C in a shaking incubator (200 rpm) for 72 h. The foaming activity was calculated according to Equation (1), [11].
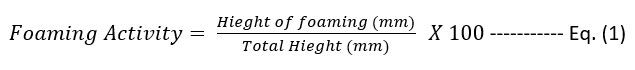
Emulsification activity
Emulsification activity was calculated by emulsification index known as E24 % according to the procedure reported by Bento et al. [17]. Emulsification assay was carried out by adding 2 ml of xylene and hexane, separately on 2 ml of each cell free supernatant of the tested bacterial isolate. Distilled water was used as control. All mixtures were mixed by vortex at high speed for 2 min, and left at room temperature for 24 h. Emulsification index E24 % was calculated according to equation (2), [17].
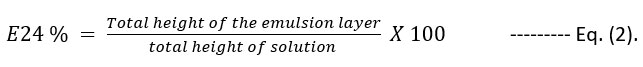
Identification of biosurfactant producing bacteria using 16S rRNA Technique
Biosurfactant producing bacterial strain was identified by 16S rRNA. Bacterium genomic DNA was extracted and cleaned up and then used as template to amplify the 16S rRNA gene by polymerase chain reaction (PCR). The universal primer 27F of (5`-AGA GTT TGA TCC TGG CTC AG-3`) and 1492R (5`-CGG YTA CCT TGT TAC GAC TT-3`) was used to amplify the 16S rRNA. The PCR reaction was run with the following thermal profile; initial denaturation for 5 min at 94 °C, denaturation for 1 min at 94 °C, annealing for 1 min at 55 °C and extension for 2 min at 72 °C and the final extension for another 1 ml of first and 10 min at 72 °C. The previous cycles were repeated for 30 times.
Determination of biosurfactant group type using rhamnose test
Rhamnose analysis was used to test the existence of carbohydrate groups in the biosurfactant molecule as stated by Dubois, et al. [18]. A volume of 0.5 ml of supernatant culture was combined with 0.5 ml of 5% phenol and 2.5 ml of sulfuric acid and incubated for 15 min. The same steps were applied on previously prepared different concentrations of rhamnose solutions (2, 4, 6 and 8 mg / L). Absorbance was measured using UV Spectrophotometer (Hach DR 6000TM, USA) at its respective λmax 490 nm. The results were plotted on absorbance curve to compare the colors of prepared concentrations with the test.
Results and Discussion
Water quality assessment studies
To evaluate the ability of a given bacterial species in a characterized community to produce biosurfactant with referring to the quality of its habitat, water quality assessment studies have been carried out for the collected water samples (Table 1). The characteristics of river Nile water at site (1) and (2) are in accordance with the international guidelines of river waters, except for oil and grease and BOD parameters of site (2) where they recorded 119 and 45 mg /L, respectively, due to the accidental pollution with oil during the ship fixation. The total bacterial count at site (2) is very high when compared with that of site (1) 15000 and 1200 CFU / mL, respectively. Such findings of site (2) are in accordance with what reported by Ewida [19] who have been assessing the water quality of river Nile during oil spills, he detected high Oil and grease, BOD values and high counts of bacteria. On the other hand, Rahawy drain wastewater (site 3) was highly contaminated (Table 1) with TSS, ammonia, BOD and bacteria, even so, the oil and grease parameter measured 7 mg /L, which is very low when compared with that of site (2).
Table-1: Water quality assessment for different sampling sites
| Water Quality Parameter | Unit | Site (1) | Site (2) | Site (3) |
| Site Description | Clear river Nile water | Oil-contaminated river Nile water | Heavily contaminated drain water | |
| pH | 7.4 | 7.8 | 8.2 | |
| Total Alkalinity | mg/ L | 124 | 136 | 360 |
| Electrical Conductivity (EC) | mmhos/cm | 0.32 | 0.37 | 1.48 |
| Total Dissolved Solids (TDS) | mg/ L | 231 | 330 | 890 |
| Total suspended Solids (TSS) | mg/ L | 10.5 | 16 | 266 |
| Turbidity | mg/ L | 4.1 | 13 | 119 |
| Ammonia | mg/ L | 0.18 | 0.29 | 14 |
| Oil & grease | mg/ L | 0.09 | 119 | 7 |
| BOD | mg/ L | 11 | 45 | 90 |
| Total Bacterial Count TBC 35 oC | CFU/ mL | 1200 | 15000 | 160000 |
| Total Coliform Count | CFU/100 mL | 2900 | 3200 | 890000 |
| Fecal Coliform Count | CFU/100 mL | 260 | 300 | 460000 |
Screening of all bacterial isolates for biosurfactant production
Table-2: Results of biosurfactant screening assay for all the isolated bacteria
|
Isolate Code |
*Blood Hemolysis | Oil
Spreading (mm) |
Drop Collapse | Foaming Activity (%) | Emulsification Activity (%) | |||
| Xylene | Hexane | |||||||
| 1 | Clear River Nile Water | CRNW1 | +ve (β+) | 30 | +ve | 33 | 11 | 10 |
| 2 | CRNW2 | -ve | -ve | -ve | 6 | 0 | 13 | |
| 3 | CRNW3 | -ve | -ve | -ve | 9 | 0 | 0 | |
| 4 | CRNW4 | +ve (α+) | 10 | +ve | 9 | 7 | 16 | |
| 5 | CRNW5 | -ve | -ve | -ve | 7 | 0 | 0 | |
| 6 | CRNW6 | -ve | -ve | -ve | 16 | 10 | 0 | |
| 7 | Oil-Contaminated Nile Water | OCNW1 | +ve (β+) | 26 | -ve | 29 | 35 | 31 |
| 8 | OCNW2 | +ve (α+) | 22 | +ve | 28 | 33 | 36 | |
| 9 | OCNW3 | -ve | -ve | -ve | 4 | 0 | 0 | |
| 10 | OCNW4 | -ve | -ve | -ve | 8 | 0 | 0 | |
| 11 | OCNW5 | -ve | 3 | -ve | 12 | 0 | 0 | |
| 12 | OCNW6 | +ve (β+) | 29 | +ve | 31 | 10 | 22 | |
| 13 | OCNW7 | +ve (α+) | 34 | -ve | 33 | 26 | 12 | |
| 14 | OCNW8 | +ve (β+) | 35 | -ve | 29 | 44 | 13 | |
| 15 | OCNW9 | +ve (α+++) | 90 | +ve | 85 | 69 | 86 | |
| 16 | El-Rahawy Drain Water | RDW1 | -ve | -ve | -ve | 13 | 0 | 11 |
| 17 | RDW2 | -ve | -ve | -ve | 11 | 0 | 0 | |
| 18 | RDW3 | -ve | -ve | -ve | 9 | 0 | 9 | |
| 19 | RDW4 | +ve (β++) | 45 | +ve | 55 | 0 | 33 | |
| 20 | RDW5 | +ve (β+) | 48 | -ve | 40 | 0 | 45 | |
| 21 | RDW6 | +ve (β+) | 36 | -ve | 39 | 33 | 11 | |
| 22 | RDW7 | +ve (α++) | 49 | +ve | 39 | 0 | 22 | |
| 23 | RDW8 | +ve (α+) | 44 | -ve | 48 | 39 | 45 | |
| 24 | RDW9 | -ve | 4 | -ve | 17 | 0 | 0 | |
| 25 | RDW10 | +ve (α+) | 9 | -ve | 41 | 0 | 19 | |
| 26 | RDW11 | -ve | -ve | -ve | 16 | 14 | 12 | |
| 27 | RDW12 | +ve (α+) | 3 | +ve | 33 | 27 | 35 | |
| 28 | RDW13 | +ve (α+) | 19 | +ve | 31 | 29 | 40 | |
Biosurfactants play an important role in oil recovery due to its ability to decrease surface tension of petroleum hydrocarbons [20, 21]. For this purpose, finding of biosurfactant producing bacteria to enhance and facilitate oil bioremediation was considered as an important aim of the present research. So, the bacterial community of each sampling site was differentiated, as a whole, 84 bacterial isolates were collected from the different samples sites. According to the cultural, morphological and biochemical reactions they were summarized to 28 isolates (6 from clear raw river Nile water, 9 from oil-contaminated river Nile water and 13 from Rahawy drain water). All the pre-identified isolates were screened for the synthesis of biosurfactants by blood hemolysis, oil spreading technique, drop collapse assay, foaming and emulsification behavior. The results were illustrated in Table (2).
Hemolytic activity as a primary method to screen biosurfactant production has been mentioned by Carrillo et al. [13]. Bernheimer and Avigad [22] reported that the biosurfactant produced by B. subtilis could lyse the red blood cells; moreover, Roy et al. [23] mentioned that the inhibition zone of hemolytic assay is directly proportional to the concentration of surfactant. As illustrated in Figure (1) the results of blood hemolysis test indicated that there are 16 isolates (57.14 %) showed positive hemolytic activity, from them 7 (25 %) isolates were β- hemolytic and 9 (32.14 %) were α-hemolytic. While the majority of bacterial strains identified as a biosurfactant producers by many authors are β- hemolytic [11, 12, and 24], there are some strains recorded as α- hemolytic, but have a great potential in biosurfactant production [14]. In the present work isolate no. 15 of code OCNW9 creates the biggest hemolytic zone (α+++) (Figure 2).
The presence of biosurfactant induces oil displacement and clear zone formation considering oil spreading assay. The zone diameter is directly proportional to the concentration and activity of biosurfactant [25, 26]. In the present study, screening of 28 isolates for oil spreading activity recorded that 64.28% of isolates showed positive oil spreading activity that known as oil displacement and isolate no. 15 of code OCNW9 recorded the biggest displacement zone diameter (90 mm). Furthermore, oil spreading ability of OCNW9 isolate was checked out using paraffin oil, the organism showed high potency to spread paraffin oil (45 mm) (Figures 2). Hemolytic activity and oil displacement of cell free extract revealed the ability of bacterial isolates to produce biosurfactant [27].
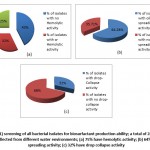 |
Figure-1: Screening of all bacterial isolates for biosurfactant production ability; a total of 28 isolates were collected from different water environments; (a) 75% have hemolytic activity; (b) 64% have oil spreading activity; (c) 32% have drop collapse activity |
 |
Figure 2: Some activities of P. protegens (a) α hemolysis (b) oil dispersing ability against vegetable oil, diameter (90 mm); (c) oil dispersing ability against paraffin oil, diameter (45 mm) |
The drop collapse assay was described by Jain et al. [16] and depended on the destabilization of liquid drop by cell free extract containing biosurfactant. The drop collapse method is rapid and easy to carry out, requiring a small volume of sample and no specialized equipment. Nine isolates (32.14 %) showed positive potential using oil collapse assay. The successful collapse test ensures the biosurfactant’s extracellular development and surface activity against oil [28]. Also, isolate no. 15 of code OCNW9 recorded positive potential against oil using oil collapse assay as shown in Table (2). The foaming ability test was carried out to the 28 isolates under investigation, according to Abouseoud et al. [29]. Isolate no 15 of code OCNW9 was revealed the highest foaming activity.
 |
Figure 3: Emulsification activity of P. protegens: (a) vegetable oil where E24 was 50%; (b) paraffin oil where E24 was 85%; (c) xylene where E24 was 60%; (d) hexane where E24 was 86% |
Emulsification ability was considered as accurate assay to screen isolates producing biosurfactant [30]. The formed emulsion was stable and the organism was considered as a good bio-surfactant producer if its E24 for many organic liquids is equal to or more than 50% [31]. Among all the studied isolates, only OCNW9 isolate could form the highest stable emulsions with xylene and hexane. Furthermore, as shown in Figure (3), emulsion activity (E24 %) of OCNW9 isolate against vegetable oil, paraffin oil, xylene and hexane separately was 50%, 85%, 69 % and 86 % respectively. Concerning the habitat of isolate no 15, code OCNW9 which showed the highest ability to produce biosurfactant, it was isolated from the raw river Nile water contaminated with some oil spills. Many researchers reported the occurrence of biosurfactant producing bacteria in hydrocarbon-contaminated environments [32-34].
Identification of best biosurfactant producing bacteria using 16S rRNA technique
From the results showed in Table (2), it is clear that among all isolates, isolate no. 15 of code OCNW9 was the best biosurfactant producer that showed the highest hemolysis, oil displacement, emulsification activity against various oils. It was selected for identification by 16S rRNA sequence analysis. The genomic DNA of OCNW9 was amplified using polymerase chain reaction (PCR) and PCR product was sequenced using 16S rRNA sequencer. The OCNW9 bacterial isolate was identified as Pseudomonas protegens with accession No. given from GenBank MN396228.
Determination of biosurfactant group type using rhamnose test
Rhamnose test was performed as mentioned by Dubios [18] the results obtained indicated that the biosurfactant produced by P. protegens in the present study belongs to rhamnolipid group (Figure 4). Concerning such results, many strains of the genus Pseudomonas were reported in the literature as biosurfactants producers [1, 29]. Furthermore, Vandana and Peter [35] isolated and identified Pseudomonas flouriscence from oil contaminated water environment and have found that the biosurfactant was belong to rhamnolipid group.
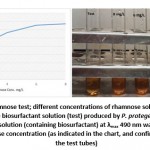 |
Figure 4: Results of rhamnose test; different concentrations of rhamnose solutions (2, 4, 6, and 8 mg/ L) were used against the biosurfactant solution (test) produced by P. protegen. The absorbance of the color given by the test solution (containing biosurfactant) at λmax 490 nm was 2 which point to more than 2 mg/ L of rhamnose concentration (as indicated in the chart, and confirmed by color intensity in the test tubes) |
Conclusion
Three different water habitats were investigated, the chemical and microbiological features, as well as, the bacterial communities were characterized. The first was clear and of normal characteristics as a river water. The second was suffering from oil contamination with high oil and grease value, while the third was wastewater with high content of chemical contaminants, but the oil and grease value is low. The bacterial community of each site was characterized and screened for biosurfactants production; the best biosurfactant producing bacterial isolate was belonging to the community of oil contaminated site. It was identified by 16s rRNA technique as Pseudomonas protegens MN396228. Such bacterium produces a biosurfactant with high ability to; i) displace vegetable oil (90%) and paraffin oil (64%), and ii) emulsify vegetable oil, paraffin oil, xylene and hexane by 50%, 85%, 60% and 86%, respectively. The identification of such biosurfactant was belonging to rhamnolipid group. The authors recommend the use of such bacterium in the field of oil bioremoval, and the biosurfactant in the field of industry.
References
- Chen S. Y., Wei Y. H., Chang J. S. Repeated pH-stat fed-batch fermentation for rhamnolipid production with indigenous Pseudomonas aeruginosa Appl Microbiol Biotechnol. 2007;76(1):67-74. http:// doi:10.1007/s00253-007-0980-2
- Gudiña E. J., Teixeira J. A., Rodrigues L. R. Biosurfactant-producing lactobacilli: screening, production profiles, and effect of medium composition. Applied and Environmental Soil Science. 2011;(2011):1 – 9. http://doi:10.1155/2011/201254
- Saharan B., Sahu R., Sharma D. A review on biosurfactants: fermentation, current developments and perspectives. Genetic Engineering and Biotechnology 2011;1:1-14.
- Anderson R., Newman M. The chemistry of the lipids of tubercle bacilli XXXIII. Isolation of trehalose from the acetone-soluble fat of the human tubercle bacillus. J Biological Chemistry. 1933;101(2):499-504.
- Desai J. D., Banat I. M. Microbial production of surfactants and their commercial potential. Mol. Biol. Rev. 1997;61(1):47-64.
- Nuñez A., Foglia T. A., Ashby R. Enzymatic synthesis of a galactopyranose sophorolipid fatty acid-ester. Biotechnology letters. 2003;25(16):1291-7.
- Thavasi R. Microbial biosurfactants: from an environmental application point of view. Bioremed Biodegrad. 2011;2(5).
- Femi-Ola T., Oluwole O., Olowomofe T., Yakubu H. Isolation and screening of biosurfactant-producing bacteria from soil contaminated with domestic waste water. 2015;3:58-63.
- Akintokun A. K., Abibu W. A., Oyatogun M. O. Microbial dynamics and biogas production during single and co-digestion of cow Dung and rice Husk. Envi. Res. 2017;39(2):67-76.
- Standard Methods for the Examination of Water and Wastewater. American Public Health Association (APHA), 23rd 2017. DOI:10.2105/SMWW.2882.193.
- Youssef N. H., Duncan K. E., Nagle D. P., Savage K. N., Knapp R. M., McInerney M. J. Comparison of methods to detect biosurfactant production by diverse microorganisms. J Microbiol Methods. 2004;56(3):339-47. http://doi:1016/j.mimet.2003.11.001
- Ibrahim M., Ijah U., Manga S., Bilbis L., Umar S. Production and partial characterization of biosurfactant produced by crude oil degrading bacteria. International Biodeterioration & Biodegradation. 2013;81:28-34. https://doi.org/10.1016/j.ibiod.2012.11.012
- Carrillo P., Mardaraz C., Pitta-Alvarez S., Giulietti A. Isolation and selection of biosurfactant-producing bacteria. World J Microbiol Biotechnol.1996;12(1):82-4. http://doi: 10.1007/BF00327807.
- Banat I., Thavasi R., Jayalakshmi S. Biosurfactants from marine bacterial isolates. Current research, technology and education topics in applied microbiology and microbial biotechnology. 2011; pp 1367-73.
- Morikawa M., Hirata Y., Imanaka T. A study on the structure–function relationship of lipopeptide biosurfactants. Biophys. Acta. 2000;1488(3):211-18. http://doi:10.1016/s1388-1981(00)00124-4
- Jain D., Collins-Thompson D., Lee H., Trevors J. A drop-collapsing test for screening surfactant-producing microorganisms. J Microbiological Methods. 1991;13(4):271-9.
- Bento F. M., de Oliveira Camargo F. A., Okeke B. C., Frankenberger W. T. Diversity of biosurfactant producing microorganisms isolated from soils contaminated with diesel oil. Microbiol Res. 2005;160(3):249-55. http://doi:1016/j.micres.2004.08.005
- Dubois M., Gilles K. A., Hamilton J. K., Rebers P. T., Smith F. Colorimetric method for determination of sugars and related substances. Analytical chemistry.1956;28(3):350-6.
- Ewida, A. Y. I. Oil Spills: Impact on water quality and microbial community on the Nile River, Egypt. j. Environ. 2014;3(4): 192-8.
- Matvyeyeva O. L., Vasylchenko A., Aliievа O. R. Microbial biosurfactants role in oil products biodegradation. International J of Environmental Bioremediation & Biodegradation. 2014;2( 2):69-74. http://doi:10.12691/ijebb-2-2-4
- Bernheimer A., Avigad L. S. Nature and properties of a cytolytic agent produced by Bacillus subtilis. Microbiology.1970;61(3):361-9. http://doi:1099/00221287-61-3-361
- Roy S., Chandni S., Das I., Karthik L., Kumar G., Rao K. V. B. Aquatic model for engine oil degradation by rhamnolipid producing Nocardiopsis VITSISB. 3. 2015;5(2):153-64. http://doi:10.1007/s13205-014-0199-8
- Anandaraj B., Thivakaran P. Isolation and production of biosurfactant producing organism from oil spilled soil. J Biosci. Tech.2010;(3):120-6.
- Huy N. Q., Jin S., Amada K., Haruki M., Huu N. B., Hang D. T., Ha D. T. C., Imanaka T., Morikawa M., Kanaya S. Characterization of petroleum-degrading bacteria from oil-contaminated sites in Vietnam. J Biosci. Bioeng.1999;88(1):100-2.
- Urum K., Pekdemir T. Evaluation of biosurfactants for crude oil contaminated soil washing. 2004;57(9):1139-50. http://doi:10.1016/j.chemosphere.2004.07.048
- Abouseoud M., Maachi R., Amrane A., Boudergua S., Nabi A. Evaluation of different carbon and nitrogen sources in production of biosurfactant by Pseudomonas fluorescens. 2008;223(1-3):143-51. https://doi.org/10.1016/j.desal.2007.01.198
- Bosch M. P., Robert M., Mercade M., Espuny M., Parra J., Guinea J. Surface active compounds on microbial cultures: investigation and production of surface active compounds on microbial cultures. Tenside Detergents. 1988;25(4):208-11.
- Yateem A., Balba M., Al-Shayji Y., Al-Awadhi N. Isolation and characterization of biosurfactant-producing bacteria from oil-contaminated soil. Soil and Sediment Contamination. 2002;11(1):41-55.
- Bodour A. A., Drees K. P., Maier R. M. Distribution of biosurfactant-producing bacteria in undisturbed and contaminated arid southwestern soils. Appl Environ Microbiol. 2003;69(6):3280-7. http://doi:1128/aem.69.6.3280-3287.2003
- Das K., Mukherjee A. K. Characterization of biochemical properties and biological activities of biosurfactants produced by Pseudomonas aeruginosa mucoid and non-mucoid strains isolated from hydrocarbon-contaminated soil samples. Appl Microbiol Biotechnol. 205;69(2):192-9. http://doi:1007/s00253-005-1975-5
- Vandana P., Peter J. K. Production, partial purification and characterization of biosurfactant produced by Pseudomonas fluorescens. International J Advanced Technology in Engineering and Science. 2014;2(7):258-64.

This work is licensed under a Creative Commons Attribution 4.0 International License.





