How to Cite | Publication History | PlumX Article Matrix
Ahmed Atef1,2
![]() , Mahmoud A. Abd El-Hafiez1
, Mahmoud A. Abd El-Hafiez1 , Fotouh M. El-Domyati1
, Fotouh M. El-Domyati1
![]() , Diana A.H. Al-Quwaie3
, Diana A.H. Al-Quwaie3 , Nouf H. Alsubhi3
, Nouf H. Alsubhi3
![]() , Aala A. Abulfaraj3
, Aala A. Abulfaraj3
![]() , Rana A. Alghamdi4
, Rana A. Alghamdi4
![]() , Ahmed Bahieldin1
, Ahmed Bahieldin1 ,2,* and Mohamed A. Rashed1
,2,* and Mohamed A. Rashed1
1Department of Genetics, Faculty of Agriculture, Ain Shams University, Cairo, Egypt
2Department of Biological Sciences, Faculty of Science, King Abdulaziz University (KAU), P.O. Box 80141, Jeddah 21589, Saudi Arabia
3Department of Biological Sciences, Science and Arts College, Rabigh Campus, King Abdulaziz University (KAU), Jeddah, Saudi Arabia
4Department of Chemistry, Science and Arts College, Rabigh Campus, King Abdulaziz University (KAU), Jeddah, Saudi Arabia
Corresponding Author E-mail : abmahmed@kau.edu.sa
DOI : http://dx.doi.org/10.13005/bbra/2837
ABSTRACT: The technology of transcription activator-like effector (TALE) is based on designing and synthesizing DNA binding protein as a molecular tool for specific targeting of any desired gene. TALE-like proteins were originally identified in bacteria including two plant pathogenic bacteria and one fungal symbiotic bacteria (namely Paraburkholderia rhizoxinica). Plant transcription activator protein can be customly designed based on the information of the original Paraburkholderia TALE-like protein (BAT). In the present study, we designed a new protein in order to reorder the repeat domain to specifically target the minimal BS3 promoter. This modified BAT sequence was fused to nuclear localization signal (NLS) at the 5` end and Herpes simplex virus VP16 activation domain at the 3` end, then designed protein sequence was reverse translated, de novo synthesized and cloned in a plant expression system. Nicotiana benthamiana plants were co-transformed via agro-infiltration method with the expression cassette with the modified BAT sequence to target the effector binding element (EBE) of the minimal BS3 promoter that drives GUS gene of the mBS3::GUS cassette. Two days post inoculation, plant leaves were GUS-stained and strong signal was detected as indicator for GUS gene activation by the modified BAT sequence. This data suggests that the synthesized BAT protein could be used as a custom transcription activator of any desired genes in plant.
KEYWORDS: BAT; EBE; GUS; TALE; Transcription Activator
Download this article as:| Copy the following to cite this article: Atef A, El-Hafiez M. A. A, El-Domyati F. M, Diana A.H. Al-Quwaie, Alsubhi N. H, Abulfaraj A. A, Alghamdi R. A, Bahieldin A, Rashed M. A. Modification of Paraburkholderia TALE-Like Protein to Activate Transcription of Target Genes in Plant. Biosci Biotech Res Asia 2020;17(2). |
| Copy the following to cite this URL: Atef A, El-Hafiez M. A. A, El-Domyati F. M, Diana A.H. Al-Quwaie, Alsubhi N. H, Abulfaraj A. A, Alghamdi R. A, Bahieldin A, Rashed M. A. Modification of Paraburkholderia TALE-Like Protein to Activate Transcription of Target Genes in Plant. Biosci Biotech Res Asia 2020;17(2). Available from: https://bit.ly/37vWNk5 |
Introduction
Many approaches to controlling gene expression were undertaken. They include over-expression of natural transcription factors (TFs) in transgenics, tissue-specific expression of transcription factors and modified expression of transcription factors. Some of these methods were successful, but none has proven to be broadly applicable as the case of synthetic transcription factors.1 Synthetic transcriptional factors are hybrid of (i) designed DNA binding domains that recognize specific sequences in the promoter present upstream the target gene; (ii) effector domains that activate or repress the transcription of target genes; and (iii) nuclear localization signal (NLS) for translocating the synthetic TF to the nucleus where transcription occurs. The protein product of the designed transcription factors is engineered to target specific genomic sequences to enable the regulation of the target genes.1
Zinc finger proteins (ZF) are first protein domain engineered to in vitro bind to any chosen locus2. Engineering of the modular DNA binding domain (DBD) of TALE protein with predetermined sequence specificity is a first step to building synthetic transcription factors. Recently, Transcriptional Activator-Like Effector (TALE) DNA binding domains from plant pathogen Xanthomonas sp have also been used to target pre-determined genomic loci.3-7 TALEs function as transcription activators during infection as one of a large group of effector proteins secreted in plant cells via type III secretion system. The N-terminal of TALE contains secretion and translocation signal. The central part of this protein contains unique DNA binding domain composed of repeating modular DNA-binding domain, nuclear localization signals (NLS) and a highly conserved acidic activation domain (AD).8 The central repetitive sequence is nearly identical except the residues 12 and 13, named repeat variable di-residues (RVDs).9 Different TALEs vary in repeat number and RVD composition.8
Many TALE homologs were identified in different bacterial species (Ralstonia solanacearum and Paraburkholderia rhizoxinica.10-12 The pathogenesis role in plants was major concern for scientists until the DNA binding code was deciphered.9,13 The latter groups identified the four major RVDs comprising HD, NI, NG and NN, which bind to cytosine (C), adenine (A), thiamine (T), and guanine (G)/A, respectively.
The major challenge of using of DNA binding domain of TALE in plants is the pathogenic nature of Xanthomonas and Ralstonia TALE, which may lead to adaptive immunity in some plant species. Testing new TALE-like protein in plant system could increase our repertories for plant gene expression custom control. The present work aimed at developing new synthetic transcription factor for controlling gene expression in plants based on nonpathogenic bacteria.
In the present study, we used Paraburkholderia TALE-like protein to synthesize new TALE from non-pathogenic origin and approach to generate this TALE can be used to monitor expression of any desired gene.
Materials and Methods
Overall description modified Paraburkholderia TALE-like protein construction to activate transcription of target gene in plants is shown in Figure 1.
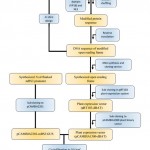 |
Figure 1: Description of the modified Paraburkholderia TALE-like protein |
Transcription Activator Designing
Paraburkholderia TALE-like protein sequence (uniprot: E5AV36) was modified by reordering repeat domain region at the DNA level to the new repeat variable di-residues (RVDs) sequence NI-NG-NI-NG-NI-NI-NI-ND-ND-NG-NI-NI that target the effector binding element (EBE) (e.g., ATATAAACCTAA) of the minimal BS3 promoter. Subsequently, DNA sequence of the nuclear localization signal (NLS) (encoding PKKKRK) was fused to N-terminus and DNA sequence of the Herpes simplex virus VP16 (encoding ADFEFEQMFTDALGIDELPQ) was fused to the DNA sequence encoding the C-terminus of the protein. Modified DNA sequence was reverse translated using Vector NTI Software v11.5 (Thermo Fisher, USA). Codon usage table of Nicotiana benthamiana [gbpln]:100 (Table 1) was used as a reference and codons for each amino acid with the highest frequencies were selected taking into consideration the avoidance of repetitive sequences. NcoI restriction site were added to reverse translated sequence by adding two cytosine bases upstream open reading frame (ORF), this addition was enough to generate NcoI site, XbaI site was added downstream ORF.
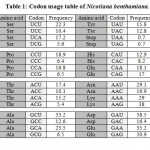 |
Table 1: Codon usage table of Nicotiana benthamiana. |
DNA Synthesis and Vectors Construction
The modified reverse translated sequence was synthesized by Integrated DNA Technology Inc. (IDT, USA) and cloned in pIDSMART. Synthesized BAT was subcloned in pRT103 plant expression vector14 at NcoI and XbaI restriction sites. HindIII fragment of pRT103 was subsequently cloned to binary vector pCAMBIA1300, 10 colonies were picked and cultivated overnight in Luria-Bertani (LB) broth media supplemented with 50 µM kanamycin, plasmids were extracted by alkaline lysis15 and digested with HindIII restriction enzyme, positive clones were termed pCAMBIA1300-dBAT (Figure 2).
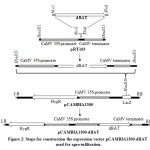 |
Figure 2: Steps for construction the expression vector pCAMBIA1300-dBAT |
Minimal BS3 promoter (P-mBS3) fragment (139 bp) flanked by NcoI restriction site (Figure 3) were synthesized and cloned to pIDSMART by Integrated DNA Technology Inc. (IDT, USA), and resulted mBS3 clones were digested with NcoI restriction enzyme followed by agarose gel electrophoresis. Then, the P-mBS3 fragment was purified by QIAquick Gel Extraction kit (Cat. No. 28704, Qiagen INC., USA) following the instruction manual. Plant binary vector pCAMBIA2201 was digested with NcoI restriction enzyme to remove tail-to-tail CaMV 35S promoters followed by electrophoresis and a 9914 bp fragment was purified from the gel. P-mBS3 NcoI fragment and the 9914 bp NcoI-digested pCAMBIA2201 fragment was ligated using T4 ligase (New England Biolabs, USA). Resultant of ligation reaction was transformed in E. coli strain MachI. Transformed bacteria were plated on LB medium supplemented with 50 µM kanamycin and 2 µg/ml X-Gal overnight of which 10 white colonies were selected for cultivation and plasmid extraction. Resulted plasmids were checked for the incorporation of P-mBS3 by NcoI digestion followed by gel electrophoresis. Then, 5 ng of each positive clone were used for polymerase chain reaction (PCR) using 10 pmol primers of OriF: CAGCCGATTGTCTGTTGTG (in the NPTII gene upstream the promoter at bases 9823 up to 9843 of pCAMBI2201) and 10 pmol of OriR: CGGGTGACAAGAACAAGAGG (within P-mBS3 at bases 10002 up to 9082 of pCAMBI2201-mBS3::GUS), 5 μL GoTaq® Master Mix, and final reaction volume adjusted to 10 μL. PCR amplification was carried out in a thermocycler programmed as the following: initial start separation cycle at 94°C for 2 min, 32 cycles including a denaturation step at 94°C for 30 sec, an annealing step at 52°C for 30 sec and polymerization step at 72°C for 30 sec, Then a final extension cycle at 72°C for 10 min. The PCR products (180 bp) were screened by agarose gel electrophoresis at 2% and positive clones, which indicate the right promoter orientation were termed pCAMBIA-mBS3 (Figure 4). Then, 1 ug DNA each from pCAMBIA-mBS3::GUS and pCAMBIA1300-dBAT plasmids were used in transforming Agrobacterium. tumefaciens GV3101. Bacterial transformation was carried out using freeze-thawing method.16
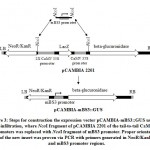 |
Figure 3: Steps for construction the expression vector pCAMBIA-mBS3::GUS |
 |
Figure 4: Minimal BS3 promoter sequence (139 bp). NcoI restriction |
Nicotiana Benthamiana Leaf Transformation and Transient Gene Expression Assay
Four-week-old N. benthamiana leaves were infiltrated by A. tumefaciens co-transformed with pCAMBIA-mBS3::GUS and pCAMBIA1300-dBAT. pCAMBIA-mBS3::GUS alone was infiltrated in N. benthamiana as a negative control. Agro-infiltration was conducted according to Leuzinger et al.17 and Li et al.18. Infiltrated 1-cm discs were collected 48 h post inoculation. Then, 4 leaf discs were stored at -80°C for RNA extraction. Then, 2 leaf discs were used for qualitative GUS assay. GUS staining was conducted by immersing leaf discs in GUS staining solution (which contains 10 mM sodium phosphate [pH 7], 10 mM EDTA, 0.1% Triton X-100, 0.1% 5-bromo-4-chloro-3-indolyl-β-D-glucuronide [X-Gluc], 1 mM potassium ferricyanide and 1 mM potassium ferrocyanide) at 37°C for 24 h. The discs were cleared in ethanol and photographed.
Validation of GUS Assay via sqRT-PCR
Total RNA was isolated from 2 leaf discs of 4 different infiltrated plants using SV Total RNA extraction kit (Promega Inc., USA) according to the manufacturer protocol. First-strand cDNA synthesis was performed by The ImProm-II™ Reverse Transcription System (Promega Inc., USA) as follows: 1.5 μg of total RNA and 50 μmol poly-T primer were incubated at 70°C for 5 min, then reaction quickly chilled on ice before addition of 2 μL of 10 mM dNTPs, 5 μL of 5x buffer, 1 μL of 40 U RNasin® Ribonuclease Inhibitor and 1 μL of ImProm-II™ Reverse Transcriptase in a total of 25 μL mixture. Then, the mixture was incubated at 42°C for 1 h and 5 μL of first strand reaction were used as template for PCR as follows: 12.5 μL GoTaq® Master Mix, 10 pmol of each primer (gusF: GCGTAATGCTCTACACCACG, gusR: AAGGGTAATGCGAGGTACGG). PCR product of bp was expected to generate. Actin gene was used as a house-keeping. For PCR, 10 pmol actnF: AAGATACTCACAGAAAGAGGCTACTC and 10 pmol actnR: GGGAGCTAATGCAGTAATTTCCTT, were used to amplify bp and reaction volume was adjusted to 25 μL by ddH2O. PCR conditions for amplifying actin gene fragment was the same as those used for amplifying GUS gene fragment.
Results and Discussion
Transcription activator like effectors (TALE) from Xanthomonas sp. is used as a modular DNA binding protein. However, Xanthomonas sp. is a plant pathogen, but defense mechanisms are developed by plants to neutralize Xanthomonas TALE.19 TALE-like proteins identified in several bacterial species, particularly the repeat domain region.20 One of those is Paraburkholderia rhizoxinica, which is an endosymbiotic bacterium of Rhizopus microspores. Paraburkholderia TALE-like protein is computationally identified from Paraburkholderia rhizoxinica genome.12
Design of Transcription Activator
Paraburkholderia TALE-like protein repetitive sequences has similarity with Xanthomonas TALE repeats except for the lack of TALE secreting signal in N-terminus and the activation domain and the nuclear localization signal (NLS) in C-terminus (Figure 5). These domains are crucial to translocation and function in plant cells as transcriptional activators8. In this study, nuclear localization signal (PKKKRK) was fused to N-terminus and Herpes simplex virus VP16 (ADFEFEQMFTDALGIDELPQ) was fused to C-terminus of the original Paraburkholderia TALE-like protein (BAT) (uniprot: E5AV36) to facilitate transcription activation function in plants (Figure 5). New modified NLS:BAT2:VP16 protein sequence (Figure 6) was reverse translated following codon usage table of Nicotiana benthamiana (Nb)[gbpln]:100 used as a reference. Nb translation table was used to maximize protein translation rate in the cell and avoid the recovery of premature or troncated proteins. NcoI and XbaI resteriction site were added at the 5´and 3´ ends, respectively, for subsequent cloning into pRT103 plant expression vector (Figure 7). Synthesized DNA sequence of the new BAT was confirmed by Sanger sequencing in which results indicated 100% match with our designed sequence (Figure 7). Resterction digestion by HindIII enzyme for pRT103 resulted in the recovery of 2728 bp. Purified HindIII flanking region was used for subcloning into pCAMBIA1300 binary vector used for transient transformation or agro-infiltration technique. Agro-ifiltration offers a roboust method for testing expression of forign DNA in plant leaves at the transient level.
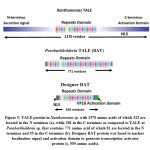 |
Figure 5: TALE protein in Xanthomonas sp. with 1370 amino acids of which 322 |
 |
Figure 6: Paraburkholderia TALE-like protein with RVD (in bold yellow letters inside |
 |
Figure 7: Designer BAT gene sequence with NcoI and XbaI restriction sites highlighted |
Transient Gene Expression Assay in Nicotiana Benthamiana Leaves
Since Paraburkholderia TALE-like protein is entirely formed of DNA binding domain (no activation domain, NLS, or secretion signal), we added Herpes simplex virus VP 16 activation domain in order to make designer transcription activator (Figure 7). We hypothesized that modification will not affect the DNA binding capability of BAT protein and the VP16 domain transcriptional activation will remain functioning. To investigate the capability of DNA binding in vivo and activate gene transcription in plants, N. benthamiana leaves were co-infiltrated by pCAMBIA1300-dBAT and pCAMBIA2201-mBS3::GUS21. Two days post inoculation, leaf discs were stained using X-Gluc stain and blue signals were detected in leaf discs co-infiltrated with the two plasmids, while no signal was detected with pCAMBIA2201-mBS3::GUS infiltrated leaves, only (Figure 8). Semi-quantitative real time polymerase chain reaction (sqRT-PCR) for GUS gene indicated that BAT transcription was detectable in N.b. plants (Figure 9).
 |
Figure 8: X-Gluc stained leaf discs indicating transcription activation of GUS |
 |
Figure 9: Semi quantitative real time PCR of actin (first row) |
Our results demonstrate that dBAT cassette is active in tobacco as DNA binding protein. Despite the previously identified TALE effector, Paraburkholderia TALE-like protein lacks NLS or transcription activation domain. Original BAT sequence failed to transport to the nucleus in plant cells22. In this study, we demonstrated adding nuclear localization signal to facilitate nuclear localization and gene activation in plant leaves in vivo via agro-infiltration technique.
Paraburkholderia TALE-like (BAT) protein is functionally different when compared to xanthomonas TALE proteins, where gene activation by dBAT results in lower signal than previously reported23. This result could be due to the lack of N-terminus and C-terminus domains, which may play important roles in protein/DNA binding stabilization. However, we suggest another strategy for enhanced synthetic transcription activator by using Paraburkholderia TALE-like repeat array with Xanthomonas TALE scaffold. This strategy may overcome the low binding capability of BAT, and also helps to escape from any defense mechanisms developed by plants such as hypersensitivity against repeat domain. Further studies are needed to test the toxicity of Paraburkholderia TALE-like protein in different plant species and to develop programmable transcription activators of controlling any desired gene with higher DNA affinity. Finally, we claim that the approach for synthesizing a new TALE of non-pathogenic origin opens an avenue towards the possibility to manipulate and monitor expression of any desired gene via transgenesis.
As compared to CRISPR/Cas9 technology that is considered an easier approach in gene edition, while TALE was abandoned as the latter is considered a time-consuming and low-efficiency process in construction controlling elements. However, CRISPR/Cas9 approach has an off-target activity that declines its application in vivo, while TALE still can be applied in vivo for gene editing and therapy because of its high targeting capability. We introduce a high-efficient method for constructing custom TALEs to overcome its key limitation. We created a new DNA binding protein as a molecular tool for specific targeting of any desired. The new method was verified by the use of GUS as a reporter gene. Our new approach is fast and less demanding. Recently, Qasim et al.24 developed a TALEN-mediated gene editing system to promote the T cell receptor α chain and CD52 gene loci. Such a bridge-to-transplantation strategy promoted the therapeutic potential of TALEN gene-editing technology.
More recently, Zhang et al.3 also used a new method to customize TALEs to be as easy and rapid as CRISPR. They developed a whole pipeline to assemble custom TALEs in as little as few hours. They verified their new method by using customized TALEs to target promoters of two transcription factor genes namely HNF4a and E47. The new constructed TALEs induced expression of two endogenous genes in two cancer cells, namely HepG2 and PANC1, which resulted in promoting successful differentiation of these cancer cells. The results of the present study as well as those of recent studies3,24 might act in re-promoting the application of TALE-based technology in more therapeutic applications in the future.
In conclusion, use of Paraburkholderia TALE-like protein in synthesizing new TALE from non-pathogenic origin successfully monitored expression of desired genes.
References
- Sera, Zinc-finger-based artificial transcription factors and their applications, Adv. Drug Deliv. Rev. 61 (2009) 513-526.
- A. Townsend, D.A. Wright, R.J. Winfrey, F. Fu, M.L. Maeder, et al., High-frequency modification of plant genes using engineered zinc-finger nucleases, Nature 459 (2009) 442-445.
- Zhang, H. Chen, J. Wang, Generate TALE/TALEN as easily and rapidly as generating CRISPR, Molecular Therapy: Methods & Clinical Development Vol. 13 (2019) 310-320.
- J. Wood, T.-W. Lo, B. Zeitler, C.S. Pickle, E.J. Ralston, et al., Targeted genome editing across species using ZFNs and TALENs, Science 333 (2011) 307.
- Huang, A. Xiao, M. Zhou, Z. Zhu, S. Lin, B. Zhang, Heritable gene targeting in zebrafish using customized TALENs, Nature Biotechnol. 29 (2011) 699-700.
- Ding, Y.K. Lee, E.A.K. Schaefer, D.T. Peters, A. Veres, et al., A TALEN genome-editing system for generating human stem cell-based disease models, Cell Stem Cell 12 (2012) 238-251.
- Li, B. Liu, M.H. Spalding, D.P. Weeks, B. Yang, High-efficiency TALEN-based gene editing produces disease-resistant rice, Nature Biotechnol. 30 (2012) 390-392.
- Boch, U. Bonas, Xanthomonas AvrBs3 Family-Type III effectors: Discovery and function, Ann. Rev. Phytopathol. 48 (2010) 419-436.
- J. Moscou, A.J. Bogdanove, A simple cipher governs DNA recognition by TAL effectors, Science 326 (2009) 1501.
- Heuer, Y.-N. Yin, Q.-Y. Xue, K. Smalla, J.-H. Guo, Repeat domain diversity of avrBs3-like genes in Ralstonia solanacearum strains and association with host preferences in the field, Appl. Environ. Microbiol.73 (2007) 4379-4384.
- Li, A. Atef, A. Piatek, Z. Ali, M. Piatek, et al., Characterization and DNA-binding specificities of Ralstonia TAL-like effectors, Mol. Plant 6 (2013) 1318-1330.
- Lackner, N. Moebius, L.P. Partida-Martinez, S. Boland, C. Hertweck, Evolution of an endofungal lifestyle: deductions from the Burkholderia rhizoxinica genome, BMC Genomics 12 (2011) 210.
- Boch, H. Scholze, S. Schornack, A. Landgraf, S. Hahn, et al., Breaking the code of DNA binding specificity of TAL-Type III effectors, Science 326 (2009) 1509-1512.
- Topfer, J. Schell, H.-H. Steinbiss, Versatile cloning vectors for transient gene expression and direct gene transfer in plants, Nucl. Acids Res. 16 (1988) 8725.
- Sambrook, D.W. Russell, Molecular Cloning, Vol. 1, 2, 3 (2001) Cold Spring Harbor Laboratory Press, 3th Edition.
- A. Wise, Z. Liu, A.N. Binns, Three methods for the introduction of foreign DNA into Agrobacterium, Methods Mol. Biol. 343 (2006) 43-53.
- Leuzinger, M. Dent, J. Hurtado, J. Stahnke, H. Lai, et al., Efficient agroinfiltration of plants for high-level transient expression of recombinant proteins, J. Vis. Exp. 77 (2013) 1-9.
- Li, R. Blue, B. Zeitler, T.L. Strange, J.R. Pearl, et al., Activation domains for controlling plant gene expression using designed transcription factors, Plant Biotechnol. J. 11 (2013) 671-680.
- Hutin, A.L. Pérez-Quintero, C. Lopez, B. Szurek, MorTAL Kombat: the story of defense against TAL effectors through loss-of-susceptibility, Front. Plant Sci. 6 (2015) 535.
- J. Bogdanove, S. Schornack, T. Lahaye, TAL effectors: Finding plant genes for disease and defense, Curr. Opin. Plant Biol. 13 (2010) 394-401.
- Römer, S. Recht, T. Lahaye, A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens, Proc. Natl. Acad. Sci. USA 106 (2009) 20526-20531.
- de Lange, C. Wolf, J. Dietze, J. Elsaesser, R. Morbitzer, T. Lahaye, Programmable DNA-binding proteins from Burkholderia provide a fresh perspective on the TALE-like repeat domain, Nucl. Acids Res. 42 (2014) 7436-7449.
- Morbitzer, P. Romer, J. Boch, T. Lahaye, Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors, Proc. Natl. Acad. Sci. USA 107 (2010) 21617-21622.
- Qasim, H. Zhan, S. Samarasinghe, S. Adams, P. Amrolia, et al., Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells, Science Translational Medicine 9 (2017) Issue 374, eaaj2013.

This work is licensed under a Creative Commons Attribution 4.0 International License.





