How to Cite | Publication History | PlumX Article Matrix
Escherichia coli in Saudi Arabia: An Overview of Antibiotic-Resistant Strains
Naiyf S. Alharbi
Department of Botany and Microbiology, College of Science, King Saud University, Riyadh 11451, Saudi Arabia
DOI : http://dx.doi.org/10.13005/bbra/2848
ABSTRACT: Escherichia coli (E. coli) bacterial strains are considered as the most important human pathogens. Health issues are increasing in complexity owing to the persistent emergence of antibiotic resistant E. coli strains, which have been isolated and detected worldwide, including Saudi Arabia. A review of the prevalent strains resistant to the standard antibiotics used in a local region is critical and may be beneficial at the national and international levels. Treatment for E. coli infection has been highly difficult by the rise of resistance to most first-line antibiotics. The present study aimed to update the scientific information regarding E. coli strains, which have the ability to resist the standard drugs used to treat bacterial infections in Saudi Arabia. The data retrieved from https://scholar.google.com and Saudi Digital Library (https://sdl.edu.sa/) indicate that E. coli strains isolated from several sources in Saudi Arabia show resistance to almost all antibiotics, except 5th generation cephalosporins (ceftraolin and ceftobiprole), which no isolate in Saudi Arabia has been recorded to resist. Based on the results of the present study, we conclude and recommend that integrated monitoring and management of the antibiotics may reduce the health risks associated with antibiotic resistant E. coli strains.
KEYWORDS: Antibiotic; Escherichia coli; Resistance; Saudi Arabia
Download this article as:| Copy the following to cite this article: Alharbi N. S. Escherichia coli in Saudi Arabia: An Overview of Antibiotic-Resistant Strains. Biosci Biotech Res Asia 2020;17(3). |
| Copy the following to cite this URL: Alharbi N. S. Escherichia coli in Saudi Arabia: An Overview of Antibiotic-Resistant Strains. Biosci Biotech Res Asia 2020;17(3). Available from: https://bit.ly/31mpHBR |
Introduction
Escherichia coli (E. coli) is a coliform rod-shaped, Gram-negative, facultative, non-spore forming bacterium of the genus Escherichia and family Enterobacteriaceae, which includes over 53 genera and 210 species (Jenkins et al., 2017; Tenaillon et al., 2010). According to a study on drug resistances it is predicted that ten million people may die from antibiotic-resistant diseases each year by 2050 if no precautions are taken to tackle the issue, among that more than three million will lose their lives to one bacterial infection by antibiotic resistant E. coli ( O’Neill J., 2016). The E. coli strains are considered as one of the few microbes that have the skill to be adapted to numerous biofunctions. These bacteria can colonize the healthy intestinal tract of several mammals including humans. They are used as an important bio-tool in several biotechnological applications. Furthermore, they have virulence factors which cause numerous diseases in humans and animals, and affect a wide range of bio-cellular processes (Kaper et al., 2004). Although E. coli strains inhabit the gastrointestinal tract of healthy humans, it is considered as one of the most pathogenic microorganisms isolated from humans. E. coli is a very versatile bacterium that can modify easily from one bio-activity to another. Highly variable mutation rates have been reported in commensal and pathogenic E. coli strains (Matic et al., 1997).
Some strains of E. coli cause infections in the urinary, intestinal, and respiratory tracts along with other diseases. The sources of pathogenic E. coli strains include contaminated water and food, and it may be transmitted through direct contact with infected people and animals or non-direct contact. Pathogenic E. coli strains may cause enteric/diarrheal illness, urinary tract diseases (UTDs) or sepsis/meningitis (Kaper et al., 2004). According to the Centers for Disease Control and Prevention (CDC) (https://www.cdc.gov/ecoli/general/index.html, December 1, 2014), pathogenic E. coli strains can be classified into six pathotypic strains as follows: Shiga toxin-producing E. coli (STEC), enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enter invasive E. coli (EIEC), and diffusely adherent E. coli (DAEC). Illnesses resulting from bacterial infections caused by pathogenic E. coli strains involve diarrhea, inflammation of the kidney (pyelonephritis), inflammation of the colon (dysentery), and hemolytic-uremic syndrome. Extreme response to such infections may lead to tissue damage, organ failure, and death (Donnenberg, 2013). These bacterial strains may develop resistance mechanisms to inhibit the effects of antibiotics and there are confirmed scientific evidences reporting that these bacterial strains can also disseminate the resistance genes to other bacteria (Morris et al., 1998).
The treatment for infectious diseases caused by E. coli strains must not include antibiotics that can replicate the risk of severe complications such as hemolytic uremia. The misdiagnosis of E. coli infection and misuse of antibiotics for treatment, may lead to the emergence of antibiotic resistant E. coli strains. A number of pathogenic and non-pathogenic E. coli strains have developed the ability to resist the standard antibiotics through numerous mechanisms, which we have discussed in this study. In Saudi Arabia, antimicrobial drug resistance genes, including β-lactam (blaSHV), gentamicin (aac(3)-IV), streptomycin ( aadA1), tetracyclines (tet(A), tet(B)), chloramphenicol (catA1,cmlA), erythromycin (ere(A)), and sulfonamide (sul1) resistance genes, have been detected among E. coli isolates (Abo-Amer et al., 2018). The present review aimed to update several scientific concepts related to antibiotic resistant E. coli strains.
Antibiotic Resistant E. coli Strains
In general, the pure bacterial isolates are considered resistant to a specific antibiotic if the minimum inhibitory concentration (MIC) (mg/L) of the antibiotics is greater than the breakpoint (mg/L). The standard assays define MIC as the lowest concentration of the antibiotic, which has the ability to inhibit the bacterial growth, using the standard methods such as two-fold macro-dilution serials, two-fold micro-dilution serials, or E-test protocol.
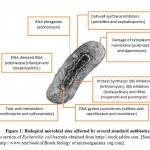 |
Figure 1: Biological microbial sites affected by several standard antibiotics. |
The MIC breakpoints are determined by several health organizations, such as the European Committee on Antimicrobial Susceptibility Test (EUCAST) and Clinical Laboratory and Standards Institute (CLSI), based on clinical and pharmacokinetic studies (Kuper et al., 2009; Reller et al., 2009). The bacterial strains, which show resistance to most standard antibiotics, are often known as superbugs. The MIC breakpoints (mg/L) and zone diameter breakpoints (mm) for most pathogenic microorganisms, including E. coli, are available in breakpoint tables for interpretation of MICs and zone diameters, Version 9.0, valid from 2019-01-01, http://www.eucast.org/clinical_breakpoints/). The breakpoints are also updated regularly by CLSI (https://clsi.org/media/2270/clsi_astnewsupdate_june2018_final.pdf). The methods used to determine MICs and MIC breakpoints must adhere to the procedures approved by the international committees on antimicrobial susceptibility testing, such as performance standards for antimicrobial susceptibility testing. 28th ed. CLSI supplement M100.
Antibiotic resistance pattern among E. coli isolates in Saudi Arabia has been evaluated using a group of standard antibiotics listed in Table 1. Figure 1 shows the percentage of antibiotics used for the treatment of urinary tract infections caused by Gram-negative bacilli, including E. coli isolates, in Buraidah Central Hospital from 1/8/2016 to 1/1/2017.
β-lactam Antibiotic Resistant E. coli Strains
Chemically, the presence of a β-lactam ring is sufficient to distinguish between the molecular structures of β-lactam antibiotics and those of the others. The β-lactam antibiotics are considered as the most widely prescribed group among all antibiotics, and include a large group of antibiotics such as penicillins (penicillin G and penicillin V, ampicillin, carbenicillin, oxacillin, piperacillin, ticarcillin , dicloxacillin, nafcillin, amoxicillin, and ampicillin), cephalosporins (cephalothin, cefoxitin, cefuroxime, ceftriaxone, cefotaxime, cefepime, ceftaroline, fosamil, and ceftolozane), monobactams (aztreonam, tigemonam, nocardicin A, and tabtoxin), carbapenems (imipenem, ertapenem, doripenem, and meropenem), and carbacephems. The β-lactam antibiotics perform a specific biological activity to inhibit bacterial cell wall biosynthesis (Demain and Elander, 1999; Elander, 2003).
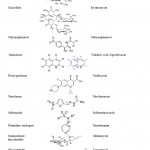 |
Table 1: Grouping of standard antibiotics used to study antibiotic resistance patterns among E. |
The β-lactam antibiotics act by penetrating the bacterial outer membrane through protein membrane channels called porins, to bind with penicillin-binding proteins (PBPs). Modifications in porins may reduce the permeability of bacterial cell membrane and β-lactam antibiotic resistance. The primary strategy followed by β-lactam antibiotic-resistant bacterial strains is the production of β-lactamase enzymes that biochemically disrupt the β-lactam ring, leading to the inactivation of the antibiotic (Bush and Bradford, 2016; Féria et al., 2002).
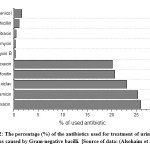 |
Figure 2: The percentage (%) of the antibiotics used for treatment of urinary tract infections caused by Gram-negative bacilli. |
Ampicillin Resistant E. coli Strains
Ampicillin (aminobenzylpenicillin) is a β-lactam and broad-spectrum antibacterial agent that can be produced from penicillin using semi-synthetic methods. Ampicillin inhibits the cell wall biosynthesis of Gram-negative and Gram–positive bacteria as well as aerobic and anaerobic bacteria. The biochemical functions of specific proteins, called PBPs, located inside the bacterial cell wall are hampered by ampicillin. Ampicillin is considered as a bacteriolytic agent, which can interfere with autolytic enzyme inhibitors such as the autolysin inhibitor. The amino group present in the chemical structure of ampicillin facilitates the passage of ampicillin through the outer membrane of pathogenic Gram-negative bacteria causing irreversible inhibition of transpeptidase enzyme, leading to bacterial lysis during the binary fission stage.
The major mechanism of resistance to β-lactam antibiotics depends on the disruption of these compounds by the β-lactamases, which destroy the amide bond of the lactam ring (Munita and Arias, 2016). Although TEM, SHV, and OXA-type β-lactamases have been detected in E. coli strains resistant to ampicillin, TEM is considered as a major β-lactamase enzyme responsible for resistance to ampicillin (Briñas et al., 2002).
Ampicillin resistant E. coli strains have been isolated and identified from patients and several environmental sources over the last 20 years in Saudi Arabia. The following table summarizes the most important findings.
Oxacillin-Resistant E. coli
Oxacillin is a β-lactam antibiotic with a narrow-spectrum activity against penicillin- and methicillin-resistant bacterial strains. It generally is described as β-lactam antibiotic resistance to penicillinase enzyme. Oxacillin may obstruct chemical transpeptidation reaction in bacterial cell walls, leading to the inhibition of peptidoglycan synthesis, which in turn causes bacterial cell autolysis (Nadarajah et al., 2006). One of the studies carried out in Saudi Arabia has reported that E. coli isolates are the most predominant among uropathogenic bacteria (N=632) and concluded that all E. coli strains have no ability to resist oxacillin (Ali, 2018).
These findings were confirmed in a study (Alharbi et al., 2018), which showed that E. coli strains (N=227) isolated from wounds did not resist oxacillin. In 2004, Shobrak and Abo-Amer reported that all E. coli strains (N=82) isolated from migratory and non-migratory wild birds were resistant strains to oxacillin (Shobrak and Abo-Amer, 2014).
Piperacillin-Resistant E. coli
Piperacillin is β-lactam antibiotic with broad-spectrum activity, classified as ureidopenicillin antibiotics, which are a class of penicillins used to treat Pseudomonas aeruginosa. Piperacillin prohibits the 3ed and final phase of the synthesis of the microbial cell wall, and is believed to inhibit autolysin inhibitors in microbial cell lysis stages. The E. coli strains can hydrolyze piperacillin via ampC and TEM-1 β-lactamase mediated in the chromosome or plasmid (Kadima and Weiner, 1997; Schechter et al., 2018). Combinations of piperacillin-tazobactam are often used to avoid these problems; nevertheless, piperacillin-tazobactam resistant E. coli strains have also been isolated and detected (Schechter et al., 2018). In Saudi Arabia, piperacillin-resistant E. coli strains have been diagnosed in clinical and non-clinical samples. Approximately 70-80% of the clinical E. coli isolates have been found to be resistant to piperacillin (Alharbi et al., 2018; Ali, 2018; Shobrak and Abo-Amer, 2014). Piperacillin-tazobactam resistant E. coli strains have been isolated from a referral hospital in Saudi Arabia, among extended-spectrum β-lactamase- and ampC β-lactamase-producing Gram-negative bacteria (Ibrahim et al., 2019).
Table 2: Percentage (%) of ampicillin resistant E. coli strains isolated from several sources in Saudi Arabia.
| Number of isolates (N) | Percentage (%) of ampicillin resistant E. coli strains. | Source of samples | Reference |
| 115 | 88.7% | Chicken | (Al‐Ghamdi et al., 1999) |
| 99 | 70.7% | Patients | (Al‐Ghamdi et al., 1999) |
| 117 | 53.8% | poultry industry workers | (Al‐Ghamdi et al., 1999) |
| 392 | In 2004 (75%)
In 2005 (80%) In 2006 (60%) In 2007 (70%) In 2008 (>80%) In 2009 (>80%) |
Patients | (Al Johani et al., 2010) |
| 50%
63% |
Outpatients
Inpatients |
(Al-Tawfiq, 2006) | |
| 37 | 78.4% | Chicken meat | (Altalhi et al., 2010) |
| 202 | 75.7% | Patients (urinary tract infection ) | (Alghoribi et al., 2015) |
| 227 | 84.8% | Patients (wound infections) | (Alharbi et al., 2018) |
| 240 | 9.1% | Raw milk | (Alharbi et al., 2018) |
| 182 | 44% | (Camel, Beef, Lamb, Poultry) | (Greeson et al., 2013) |
| 150 | 51% | Chickens | (Abo-Amer et al., 2018) |
| 683 | 85% | Patients | (Ali, 2018) |
| 157 | 83 | Outpatient | (Al Wutayd et al., 2018) |
| 82 | 70% (migratory birds)
40% (non- migratory birds) |
Birds | (Shobrak and Abo-Amer, 2014) |
Imipenem-Resistant E. coli
Imipenem (C12H17N3O4S) is one of the β-lactam carbapenem antibiotics with the ability to resist β-lactamase. It has wide spectrum activity against aerobic, anaerobic, Gram-positive, and Gram-negative pathogenic bacteria. It can be used as a combination (imipenem-cilastatin) or triple-antibiotic (imipenem-cilastatin-relebactam) product. It is reported that bacterial resistance to carbapenem antimicrobial agents (imipenem and meropenem) leads to limiting therapeutic choices.
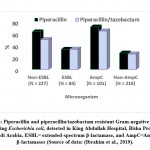 |
Figure 3: Piperacillin and piperacillin/tazobactam resistant Gram-negative bacteria |
There are generally two methods employed by E. coli strains to resist carbapenem antibiotics; producing β-lactamase enzyme or reducing the permeability of bacterial cells, and strains may sometimes use both these ways.
Resistance of E. coli strains to imipenem has been not detected in clinical and nonclinical samples in Saudi Araba (Alam et al., 2017; Alqasim et al., 2018; Saeed et al., 2018). Conversely, imipenem-resistant E. coli strains have been isolated and detected from patients (N=72) (Ali, 2018) with UTIs (N=189) (Ali, 2018) and wound infections (N=161) (Alharbi et al., 2018).
Cephalosporin-resistant E. coli
Cephalosporins are antibacterial agents classified as bactericidal β-lactam drugs. They show biological activity to inhibit the bacterial cell synthesis by blocking cell wall enzymes. Currently, there are five generations of cephalosporins, which have been produced and marketed worldwide (Shahid et al., 2009). Previous studies have reported that E. coli strains isolated from Saudi Arabia resist the first, second, third, and fourth generation of cephalosporins (i.e., cephalothin, cefoxitin, cefuroxime, ceftazidime, ceftriaxone, cefotaxime, and cefepime) (Alharbi et al., 2018; Ali, 2018). Ceftobiprole (fifth generation of cephalosporins) has been approved to treat pneumonia infections in several countries including Saudi Arabia (Pfaller et al., 2019). No evidence could be traced regarding isolation of E. coli strains resistant to ceftaroline and ceftobiprole, according to the information obtained by searching Google Scholar(https://scholar.google.com/) and (https://sdl.edu.sa/).
Tetracycline Resistant E. coli Strains
Tetracycline (C22H24N2O8), also known as anhydrotetracycline or deschlorobiomycin, is a bacteriostatic broad spectrum antibiotic that can act against an extensive range of pathogenic microbes including Gram-positive and Gram negative bacteria, chlamydiae, mycoplasmas, rickettsiae, and protozoan parasites (Chopra and Roberts, 2001). It is secondary metabolic products follows the polyketide antibiotics produced by some of the actinomycetes bacteria “Streptomyces spp.”. In general, the tetracyclines can inhibit the biosynthesis of bacterial proteins by preventing the combination of aminoacyl-tRNA with the acceptor site, in the bacterial ribosome. The biological activity of tetracycline may include the 30S, 50S bacterial ribosomal subunit, and the cytoplasmic membrane.
Table 3: Percentage (%) of Cephalosporin-resistant E. coli strains isolated from several sources in Saudi Arabia.
| Year | Cephalothin | Cefoxitin | Cefuroxime | Ceftazidime | Ceftriaxone | Cefotaxime | Cefepime | Sources | Reference |
| 2013 | – | 4 | 3 | 13 | – | 3 | 6 | Healthy neonates | (Elkersh et al., 2015) |
| 2015 | – | 15.25 | – | 81.36 | – | 76.27 | 75.58 | Patients | (Alyamani et al., 2017) |
| 2015 | – | 0 | – | 32.2 | – | 92.2 | 50 | Patients | (Al-Mijalli, 2016) |
| 2016 | 91 | 15 | 0 | 36 | 0 | 0 | 20 | Patients | (Ali, 2018) |
| 2017 | 69 | 24 | 52 | 33 | 43 | 100 | 27 | Patients | (Ali, 2018) |
| 2017 | 0 | 19 | 51.7 | 10.1 | – | 9 | 47.8 | Patients | (Alharbi et al., 2018) |
| 2017 | 0 | 0 | 8.4 | 0 | – | 0 | 0 | Raw milk | (Alharbi et al., 2018) |
| 2018 | 2 | 1 | 1 | 1 | 1 | 0 | 1 | Farm chicken | (Abo-Amer et al., 2018) |
| 2018 | 85.7 | – | 92.9 | 100 | 92.9 | 78.57 | 85.71 | Patients | (Ineta et al., 2018) |
The outer membrane of Gram-negative bacteria is traversed by the tetracyclines through the OmpF and OmpC porin pathways (Chopra et al., 1992). The bacteria can resist the tetracyclines by exporting tetracycline from the bacterial cell by efflux proteins, which are encoded by the tet efflux genes, protection of bacterial ribosomes by cytoplasmic proteins, or inactivation of tetracycline through enzymatic modification (Ref.). The misuse of tetracycline compounds has been confirmed in Saudi Arabia. Several poultry products have been screened to detect the residues of tetracycline agents, the results indicate that the level of the tetracycline residues has reached over the maximum residue limit in some tested samples (Al-Ghamdi et al., 2000).
Table 4: Percentage (%) of tetracycline-resistant E. coli strains isolated from several sources in Saudi Arabia.
| Number of isolates (N) | Percentage (%) of ampicillin resistant E. coli strains. | Source of samples | Reference |
| 116 | 99% | Chicken | (Al‐Ghamdi et al., 1999) |
| 99 | 64.7% | Patients | (Al‐Ghamdi et al., 1999) |
| 10 | 30 | Pigeons | (Abulreesh, 2011) |
| 150 | 97% | Chicken | (Abo-Amer et al., 2018) |
| 100 | 49% | inpatients (urine samples) | (Alqasim et al., 2018) |
| 683 | 85% | Patients | (Ali, 2018) |
| 161 | 68% | Patients (wound infection) | (Alharbi et al., 2018) |
| 32 | 100% (migratory birds)
84% (non- migratory birds) |
Birds | (Shobrak and Abo-Amer, 2014) |
Aminoglycosides Resistant E. coli
The aminoglycosides are natural (gentamicin and tobramycin) or semisynthetic (derivatives of natural antibiotics such as amikacin), broad-spectrum, bactericidal antimicrobial agents which are generally introduced for the treatment of Gram-negative infections in humans (Germovsek et al., 2017; Krause et al., 2016). Aminoglycoside antibiotic-resistant bacterial strains can fight the antibiotics derived from aminoglycosides, using various strategies including modification of target sites by biosynthesis of specific enzymes, as well as mutation in bacterial chromosome and efflux pump (Krause et al., 2016; Rosenberg et al., 2000). Aminoglycoside antibiotic-resistant E. coli strains have been detected in patients, individuals, and food products in Saudi Arabia (Al‐Ghamdi et al., 1999; Alharbi et al., 2018; Ali, 2018).
Gentamicin
Gentamicin (C21H43N5O7) is a secondary metabolite produced by Micromonospora purpurea (a saprophytic, filamentous, aerobic, spore-forming, and Gram-positive bacterium, which can be isolated from the soil). In general, it is used for the treatment of bacterial infections caused by bacterial strains susceptible to antibiotics, including E. coli strains. Gentamicin is classified as an aminoglycoside antimicrobial agent with broad-spectrum activity against Gram-positive and Gram-negative bacteria. It can obstruct the synthesis of bacterial proteins through interaction with the prokaryotic 30S ribosomal subunit, leading to misinterpretation of transfer ribonucleic acid (t-RNA) (Yoshizawa et al., 1998). In general, the bacteria acquire aminoglycoside resistance via three potential mechanisms: 1) alteration in bacterial cell permeability (reducing uptake), 2) modification at sites in the ribosomal subunit, and 3) synthesis of specific enzymes having the ability to modify the chemical structure of aminoglycosides (Yoshizawa et al., 1998). Aminoglycoside N(3)-acetyltransferase (aac(3)-IV) gene has been detected in numerous aminoglycoside-resistant E. coli isolates (Costa et al., 2008; Domínguez et al., 2002; Zhang et al., 2009).
Streptomycin
Streptomycin (C21H39N7O12) is chemically classified as an aminoglycoside antimicrobial agent that can be produced by Streptomyces griseus, which is frequently isolated from the soil. Streptomycin has the ability to irreversibly bind to the 30S ribosomal subunit proteins and 16S rRNA. The interaction between streptomycin and decoding area in 16S rRNA of the 30S ribosomal subunit (site near nucleotide 1400). The principle of interaction is the capacity of streptomycin to bind with a single amino acid of the 30S ribosomal protein S12 and four nucleotides of 16S rRNA, which lead to mRNA misreading. In bacteria, genetically acquired streptomycin resistance is frequently due to genetic alteration in rpsL gene, which encodes the ribosomal protein S12 (Springer et al., 2001).
In Saudi Arabia, E. coli strains resistant to gentamicin have been identified and isolated from inpatients, outpatients, animals, and foods. Spontaneous streptomycin‐resistant E coli strains have a genetic alteration in several sites in 30S ribosomal protein S12 including Lys42, Lys87, Pro90, and Gly9 (Chumpolkulwong et al., 2004). In Saudi Arabia, the E coli strains resistant to streptomycin have been characterized and streptomycin-resistance genes have been detected (Abo-Amer et al., 2018). Streptomycin-resistant E. coli strains have been isolated from numerous sources including raw chicken meat, wastewater, as well as in patient samples (skin, blood, urine, stool, and respiratory tract) (Alam et al., 2017; Altalhi et al., 2010; Mantilla-Calderon et al., 2016). In a prior study performed in Taif, Saudi Arabia, more than 48% of the streptomycin-resistant E. coli strains (N=119) were isolated from retail raw chicken meat (Al Johani et al., 2010).
Table 5: Percentage (%) of gentamicin-resistant E. coli strains isolated from several sources in Saudi Arabia.
| Number of isolates (N) | Percentage (%) of gentamicin resistant E. coli strains. | Source of samples | Reference |
| 116 | 89.7 | Chicken | (Al‐Ghamdi et al., 1999) |
| 96 | 21.9 | Patient | (Al‐Ghamdi et al., 1999) |
| 768 | 47%
(from 2006 to 2010) |
Hospitalized patient and outpatient | (Somily et al., 2014) |
| 683 | 27% | Patients | (Ali, 2018) |
| 157 | 14 | Outpatient | (Al Wutayd et al., 2018) |
| 32 | 0% (migratory birds)
4% (non- migratory birds) |
Birds | (Shobrak and Abo-Amer, 2014) |
In Riyadh, Saudi Arabia, it has been reported that all isolates of E. coli (N=200) detected in the feces of broiler chickens are resistant to streptomycin (Al-Arfaj et al., 2015). Streptomycin-resistant strains have been found to be the preponderant strains among enterotoxigenic E. coli isolates (N=181) collected from patients with diarrhea (Willshaw et al., 1995).
Tobramycin
Tobramycin (C18H37N5O9) is a narrow spectrum aminoglycoside antimicrobial agent, which can interact with microbial 30S and 50S ribosome, thereby preventing the formation of 70S ribosome complex. It is widely used to treat microbial infections caused by Gram-negative bacteria. The intracellular concentration of tobramycin is critical for its action. Active transport of tobramycin through the bacterial membrane is a significant mechanism that helps to increase tobramycin concentration inside the bacterial cell. The bacterial strains generally gain resistance to tobramycin through one or more of the three strategies mentioned above (physiological or genetic alteration in cell permeability, mutation at the ribosomal binding sites, or synthesis of enzymes having the ability to modify the aminoglycoside) (Islam et al., 2009). In Saudi Arabia, 67% of the E. coli strains (N=1116) isolated from patients were tobramycin-resistant strains (Kader and Kumar, 2004). In 2015, 57% of E. coli strains (N=130) detected in pilgrims (patients) admitted in Makkah, Saudi Arabia were tobramycin-resistant (Haseeb et al., 2016). In King Fahd Hospital University, Clinical Microbiology Department, Al-Khobar, Saudi Arabia (Al-Zahrani and Akhtar, 2005), more than 75% of the E. coli strains (N=48) depicted the ability to resist tobramycin.
Kanamycin
Kanamycin (C18H36N4O11) is a bactericidal antimicrobial agent grouped on the basis of its chemical structure in the aminoglycoside antibiotics group. It has the ability to eliminate bacterial pathogens by inhibiting protein synthesis, using the same mechanism of action as the aminoglycosides to cause irreversible damage in small ribosomal subunit and 16S ribosomal RNA. The pathogenic bacteria resistance to kanamycin. In industrial microbiology, kanamycin is produced using Streptomyces kanamyceticus. The E. coli strains with kanMX marker show resistance to kanamycin. In Kanamycin-resistant E. coli strains, efflux pumps may act to drive out kanamycin from E. coli cells. Resistance may be developed by a mutation in the ribosomal subunit target or by ribosome methyltransferases, which have gained increasing clinical importance (Garneau-Tsodikova and Labby, 2016). In Saudi Arabia, kanamycin-resistant E. coli strains have been isolated and detected in wastewater (Mantilla-Calderon et al., 2016), vegetable salads (Khiyami et al., 2011), and meat (Greeson et al., 2013). A study reported that all E. coli strains (N=60) isolated from frozen fish in Eastern Province of Saudi Arabia were kanamycin-susceptible E. coli isolates.
Neomycin
Neomycin is one of the aminoglycoside antimicrobial agents that have strong biological activity against pathogenic Gram-negative bacteria. It can be produced by fermentation using Streptomyces spp. such as S. fradiae and S. albogriseus. Neomycin inhibits microbial protein synthesis by interacting with 30S subunit and 16S rRNA. The E. coli strains that harbor the gene neo (coding for the 29-kDa phosphotransferase enzyme), have the biological ability to resist neomycin and kanamycin (Genilloud et al., 1988).
The neomycin-resistant E. coli mutants show significant alteration in the activity of membrane Mg2+-ATPase and periplasmic alkaline phosphatase. Point mutations in rrsB 16S rRNA gene, especially at the 3’ minor domain of helix 4 can lead to emergence of E. coli strains resistant to neomycin (Obaseiki-Ebor and Breeze, 1984) (https://card.mcmaster.ca/ontology/39986). In Saudi Arabia, E. coli strains resistant to neomycin have been reported by some previous studies (21% of 180 isolates of E. coli were resistant) (Abo-Amer et al., 2018); however, many reports have confirmed that all E. coli strains were susceptible to neomycin (Ali, 2018). It has been reported that neomycin is one of the less-used drugs among the 44 antibiotic drugs used to treat urinary tract infections (N=339) (Alsohaim et al., 2019).
Other Antibiotics
In Saudi Arabia, it has been reported that there are numerous E. coli strains that possess the ability to resist macrolides (erythromycin), chloramphenicol, quinolone (naldixic acid and ciprofloxacin), fluoroquinolones (norflaxacin), sulfonamide (sulfamethaxazole), glycopeptide (vancomycin), semisynthetic lincosamide (clindamycin), nitrofuran (nitrofurantoin), and pyrimidine (trimethoprim) antibiotics. Chloramphenicol, kanamycin, cefoxitin, and ceftiofur-resistant E. coli strains have been detected in Saudi Arabia from several sources of locally marketed meat (Greeson et al., 2013).
Both extended spectrum β-lactamase (ESB) E. coli or non-ESB E. coli strains show resistance to synergic action produced from sulfamethoxazole and trimethoprim (cotrimoxazole) among clinical isolates (Al-Otaibi and Bukhari, 2013). In fact, it is believed that the resistance to antibiotics is increasing continuously; however, sometimes the opposite occurs and the strains show susceptibility to the same antibiotics to which they were resistant in the past (Table).
Table 6: Percentage (%) of E. coli strains resistant to antibiotics, causing urinary tract infections in Saudi Arabia, in 1985 and 2017.
| Antibiotic | % in 2017 | Antibiotic | % in 1985 |
| Cotrimoxazole | 72 | Trimethoprim | 59 |
| Chlormphenicol | 75 | Sulfamethoxazole | 87 |
| Nalidixic acid | 76 | Nalidixic acid | 10 |
| Nitrofurantoin | 26 | Nitrofurantoin | 32 |
| Cip/Norfloxacin | 59 | N.T | – |
| Erythromycin | 100 | N.T | – |
| Clindamycin | 100 | N.T | – |
* References: (Ali, 2018); (Eltahawy and Khalaf, 1988); N.T= Not tested
Conclusion
Antibiotics are frequently used for therapy of infected humans and animals. Treatment for E. coli infection has been highly difficult by the rise of resistance to most first-line antibiotics. The data showed the prevalence of E. coli strains in Saudi Arabia, which can resist all antibiotic groups including β-lactams, cephalosporins, aminoglycosides (except fifth generation), macrolides, chloramphenicol, quinolones, flouroquiolones, nitrofurans, sulfonaides, pyrimidine analogues, semisynthetic lincosamides, glycopeptides, and tetracycline antibiotics. The integrated monitoring and management of the antibiotics used to treat infections caused by E. coli must be applied to reduce health hazards.
Acknowledgments
The author wishes to thank Dr. Jamal M. Khaled and Dr. Shine Kadaikunnan for the helpful discussions on the topic. This project was funded by the King Abdulaziz City for Science and Technology (KACST), Kingdom of Saudi Arabia (Award number: LGP- 36-7).
Conflict of Interest
There is no Conflict of interest
Funding Source
This project was funded by the King Abdulaziz City for Science and Technology (KACST), Kingdom of Saudi Arabia (Award number: LGP- 36-7).
References
- Abo-Amer, A.E., Shobrak, M.Y., Altalhi, A.D., 2018. Isolation and antimicrobial resistance of Escherichia coli isolated from farm chickens in Taif, Saudi Arabia. Journal of Global Antimicrobial Resistance 15, 65-68.
CrossRef - Abulreesh, H.H., 2011. Free living rock pigeon (Columba livia) as an environmental reservoir of enteric bacterial pathogens resistant to antimicrobial drugs in Saudi Arabia. Current Research in Bacteriology 4, 28-33.
CrossRef - Al-Arfaj, A., Hessain, A., Hemeg, H., Mohamed, K.F., Moussa, I., 2015. Isolation, characterization and antibiogram of pathogenic Escherichia coli recovered from broiler chicken, Riyadh, Saudi Arabia. J Anim Plant Sci 25, 1540-1542.
- Al-Ghamdi, M., Al-Mustafa, Z., El-Morsy, F., Al-Faky, A., Haider, I., Essa, H., 2000. Residues of tetracycline compounds in poultry products in the eastern province of Saudi Arabia. Public Health 114, 300-304.
CrossRef - Al-Mijalli, S., 2016. Extended-spectrum β-lactamase enzymes (ESBLs) produced by Escherichia coli urinary pathogens at Riyadh, Saudi Arabia. J Antimicrob Agents 2, 2472-1212.1000125.
CrossRef - Al-Otaibi, F.E., Bukhari, E.E., 2013. Clinical and laboratory profiles of urinary tract infections caused by extendedspectrum beta-lactamase-producing Escherichia coli in a tertiary care center in central Saudi Arabia. Saudi Med J 34, 171-176.
- Al-Tawfiq, J.A., 2006. Increasing antibiotic resistance among isolates of Escherichia coli recovered from inpatients and outpatients in a Saudi Arabian hospital. Infection Control & Hospital Epidemiology 27, 748-753.
CrossRef - Al-Zahrani, A.J., Akhtar, N., 2005. Susceptibility patterns of extended spectrum ß-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae isolated in a teaching hospital. Pak J Med Res 44, 64-67.
- Al‐Ghamdi, M.S., El‐Morsy, F., Al‐Mustafa, Z.H., Al‐Ramadhan, M., Hanif, M., 1999. Antibiotic resistance of Escherichia coli isolated from poultry workers, patients and chicken in the eastern province of Saudi Arabia. Tropical Medicine & International Health 4, 278-283.
CrossRef - Al Johani, S.M., Akhter, J., Balkhy, H., El-Saed, A., Younan, M., Memish, Z., 2010. Prevalence of antimicrobial resistance among gram-negative isolates in an adult intensive care unit at a tertiary care center in Saudi Arabia. Annals of Saudi medicine 30, 364.
CrossRef - Al Wutayd, O., Al Nafeesah, A., Adam, I., Babikir, I., 2018. The antibiotic susceptibility patterns of uropathogens isolated in Qassim, Saudi Arabia. The Journal of Infection in Developing Countries 12, 946-952.
CrossRef - Alam, M.Z., Alam, Q., Jiman-Fatani, A.A., Shukri, H.A., Haque, A., 2017. A surveillance study on the prevalence and antimicrobial resistance pattern among different groups of bacteria isolated from Western province of Saudi Arabia. Biomedical Research (0970-938X)
- Alghoribi, M.F., Gibreel, T.M., Farnham, G., Al Johani, S.M., Balkhy, H.H., Upton, M., 2015. Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. Journal of Antimicrobial Chemotherapy 70, 2757-2762.
CrossRef - Alharbi, N.S., Khaled, J.M., Kadaikunnan, S., Alobaidi, A.S., Sharafaddin, A.H., Alyahya, S.A., Almanaa, T.N., Alsughayier, M.A., Shehu, M.R., 2018. Prevalence of Escherichia coli strains resistance to antibiotics in wound infections and raw milk. Saudi Journal of Biological Sciences.
CrossRef - Ali, A., 2018. Antimicrobial Resistance Pattern of Uropathogens Isolated from Rafha Central Hospital, Rafha, Kingdom of Saudi Arabia. Journal of Pure and Applied Microbiology 12, 577-586.
CrossRef - Alqasim, A., Abu Jaffal, A., Alyousef, A.A., 2018. Prevalence of multidrug resistance and extended-spectrum β-lactamase carriage of clinical uropathogenic Escherichia coli isolates in Riyadh, Saudi Arabia. International Journal of Microbiology
CrossRef - Alsohaim, S.I., Bawadikji, A.A., Elkalmi, R., Mahmud, M.I.A.-d.M., Hassali, M.A., 2019. Relationship between antimicrobial prescribing and antimicrobial resistance among UTI patients at Buraidah Central Hospital, Saudi Arabia. Journal of Pharmacy & Bioallied Sciences 11, 162.
CrossRef - Altalhi, A.D., Gherbawy, Y.A., Hassan, S.A., 2010. Antibiotic resistance in Escherichia coli isolated from retail raw chicken meat in Taif, Saudi Arabia. Foodborne Pathogens and Disease 7, 281-285.
CrossRef - Alyamani, E.J., Khiyami, A.M., Booq, R.Y., Majrashi, M.A., Bahwerth, F.S., Rechkina, E., 2017. The occurrence of ESBL-producing Escherichia coli carrying aminoglycoside resistance genes in urinary tract infections in Saudi Arabia. Annals of Clinical Microbiology and Antimicrobials 16, 1.
CrossRef - Briñas, L., Zarazaga, M., Sáenz, Y., Ruiz-Larrea, F., Torres, C., 2002. β-Lactamases in ampicillin-resistant Escherichia coli isolates from foods, humans, and healthy animals. Antimicrobial Agents and Chemotherapy 46, 3156-3163.
CrossRef - Bush, K., Bradford, P.A., 2016. β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harbor perspectives in medicine 6, a025247.
CrossRef - Chopra, I., Hawkey, P., Hinton, M., 1992. Tetracyclines, molecular and clinical aspects. Journal of Antimicrobial Chemotherapy 29, 245-277.
CrossRef - Chopra, I., Roberts, M., 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232-260.
CrossRef - Chumpolkulwong, N., Hori‐Takemoto, C., Hosaka, T., Inaoka, T., Kigawa, T., Shirouzu, M., Ochi, K., Yokoyama, S., 2004. Effects of Escherichia coli ribosomal protein S12 mutations on cell‐free protein synthesis. European Journal of Biochemistry 271, 1127-1134.
CrossRef - Costa, D., Poeta, P., Sáenz, Y., Vinué, L., Coelho, A.C., Matos, M., Rojo-Bezares, B., Rodrigues, J., Torres, C., 2008. Mechanisms of antibiotic resistance in Escherichia coli isolates recovered from wild animals. Microbial Drug Resistance 14, 71-77.
CrossRef - Demain, A.L., Elander, R.P., 1999. The β-lactam antibiotics: past, present, and future. Antonie Van Leeuwenhoek 75, 5-19.
CrossRef - Domínguez, E., Zarazaga, M., Sáenz, Y., Briñas, L., Torres, C., 2002. Mechanisms of antibiotic resistance in Escherichia coli isolates obtained from healthy children in Spain. Microbial Drug Resistance 8, 321-327.
CrossRef - Donnenberg, M., 2013. Introduction In: Escherichia coli: pathotypes and principles of pathogenesis, Second Edition. Elsevier Inc. UK, Page xvii. , xvii. p.
- Elander, R., 2003. Industrial production of β-lactam antibiotics. Applied Microbiology and Biotechnology 61, 385-392.
CrossRef - Elkersh, T., Marie, M.A., Al-Sheikh, Y.A., Bloushy, A.A., Al-Agamy, M.H., 2015. Prevalence of fecal carriage of extended-spectrum-and metallo-β-lactamase-producing gram-negative bacteria among neonates born in a hospital setting in central Saudi Arabia. Annals of Saudi medicine 35, 240-247.
CrossRef - Eltahawy, A.T., Khalaf, R.M., 1988. Urinary tract infection at a university hospital in Saudi Arabia: Incidence, microbiology, and antimicrobial susceptibility. Annals of Saudi Medicine 8, 261-266.
CrossRef - Féria, C., Ferreira, E., Correia, J.D., Gonçalves, J., Caniça, M., 2002. Patterns and mechanisms of resistance to β-lactams and β-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. Journal of Antimicrobial Chemotherapy 49, 77-85.
CrossRef - Garneau-Tsodikova, S., Labby, K.J., 2016. Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. MedChemComm 7, 11-27.
CrossRef - Genilloud, O., Blázquez, J., Mazodier, P., Moreno, F., 1988. A clinical isolate of transposon Tn5 expressing streptomycin resistance in Escherichia coli. Journal of Bacteriology 170, 1275-1278.
CrossRef - Germovsek, E., Barker, C.I., Sharland, M., 2017. What do I need to know about aminoglycoside antibiotics? Archives of Disease in Childhood-Education and Practice 102, 89-93.
CrossRef - Greeson, K., Suliman, G.M., Sami, A., Alowaimer, A., Koohmaraie, M., 2013. Frequency of antibiotic resistant Salmonella, Escherichia coli, Enterococcus, and Staphylococcus aureus in meat in Saudi Arabia. African Journal of Microbiology Research 7, 309-316.
CrossRef - Haseeb, A., Faidah, H.S., Bakhsh, A.R., Al Malki, W.H., Elrggal, M.E., Saleem, F., ur Rahman, S., Khan, T.M., Hassali, M.A., 2016. Antimicrobial resistance among pilgrims: a retrospective study from two hospitals in Makkah, Saudi Arabia. International Journal of Infectious Diseases 47, 92-94.
CrossRef - Ibrahim, M.E., Abbas, M., Al-Shahrai, A.M., Elamin, B.K., 2019. Phenotypic characterization and antibiotic resistance patterns of extended-spectrum β-lactamase and AmpC β-lactamase-producing Gram-negative bacteria in a referral hospital, Saudi Arabia. Canadian Journal of Infectious Diseases and Medical Microbiology
CrossRef - Ineta, B.-E.L., Madu, E.P., Abdulhadi, A.-S.A., Ibrahim, H.I.M., 2018. Antibiotic susceptibility and plasmid profile of clinical isolates of Escherichia coli.
- Islam, S., Oh, H., Jalal, S., Karpati, F., Ciofu, O., Høiby, N., Wretlind, B., 2009. Chromosomal mechanisms of aminoglycoside resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Clinical Microbiology and Infection 15, 60-66.
CrossRef - Jenkins, C., Rentenaar, R.J., Landraud, L., Brisse, S. 2017. Enterobacteriaceae, In: Infectious Diseases. Elsevier, 1565-1578. e1562.
CrossRef - Kader, A.A., Kumar, A.K., 2004. Prevalence of extended spectrum beta-lactamase among multidrug resistant gram-negative isolates from a general hospital in Saudi Arabia. Saudi Medical Journal 25, 570-574.
- Kadima, T.A., Weiner, J.H., 1997. Mechanism of suppression of piperacillin resistance in enterobacteria by tazobactam. Antimicrobial agents and chemotherapy 41, 2177-2183.
CrossRef - Kaper, J.B., Nataro, J.P., Mobley, H.L., 2004. Pathogenic Escherichia coli. Nature Reviews Microbiology 2, 123.
CrossRef - Khiyami, M., Noura, A.-F., Busaeed, B., Sher, H., 2011. Food borne pathogen contamination in minimally processed vegetable salads in Riyadh, Saudi Arabia. Journal of Medicinal Plants Research 5, 444-451.
- Krause, K.M., Serio, A.W., Kane, T.R., Connolly, L.E., 2016. Aminoglycosides: an overview. Cold Spring Harbor Perspectives In Medicine 6, a027029.
CrossRef - Kuper, K.M., Boles, D.M., Mohr, J.F., Wanger, A., 2009. Antimicrobial susceptibility testing: a primer for clinicians. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy 29, 1326-1343.
CrossRef - Mantilla-Calderon, D., Jumat, M.R., Wang, T., Ganesan, P., Al-Jassim, N., Hong, P.-Y., 2016. Isolation and characterization of NDM-positive Escherichia coli from municipal wastewater in Jeddah, Saudi Arabia. Antimicrobial Agents and Chemotherapy 60, 5223-5231.
CrossRef - Matic, I., Radman, M., Taddei, F., Picard, B., Doit, C., Bingen, E., Denamur, E., Elion, J., 1997. Highly variable mutation rates in commensal and pathogenic Escherichia coli. Science 277, 1833-1834.
CrossRef - Morris, A., Kellner, J.D., Low, D.E., 1998. The superbugs: evolution, dissemination and fitness. Current Opinion in Microbiology 1, 524-529.
CrossRef - Munita, J.M., Arias, C.A., 2016. Mechanisms of antibiotic resistance. Microbiology Spectrum
CrossRef - Nadarajah, J., Lee, M.J., Louie, L., Jacob, L., Simor, A.E., Louie, M., McGavin, M.J., 2006. Identification of different clonal complexes and diverse amino acid substitutions in penicillin-binding protein 2 (PBP2) associated with borderline oxacillin resistance in Canadian Staphylococcus aureus isolates. Journal of Medical Microbiology 55, 1675-1683.
CrossRef - Obaseiki-Ebor, E., Breeze, A., 1984. Gentamicin-and neomycin-resistant mutants of Escherichia coli K-12 cross-resistant to nitrofurans. Journal of Antimicrobial Chemotherapy 13, 567-576.
CrossRef - O’Neill J., 2016 Tackling drug‐resistant infections globally: final report and recommendations.
- Review on Antimicrobial Resistance.
- Pfaller, M.A., Flamm, R.K., Duncan, L.R., Shortridge, D., Smart, J.I., Hamed, K.A., Mendes, R.E., Sader, H.S., 2019. Ceftobiprole activity when tested against contemporary bacteria causing bloodstream infections in the United States (2016–2017). Diagnostic Microbiology and Infectious Disease.
CrossRef - Reller, L.B., Weinstein, M., Jorgensen, J.H., Ferraro, M.J., 2009. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clinical Infectious Diseases 49, 1749-1755.
CrossRef - Rosenberg, E.Y., Ma, D., Nikaido, H., 2000. AcrD of Escherichia coli is an aminoglycoside efflux pump. Journal of Bacteriology 182, 1754-1756.
CrossRef - Saeed, W.M., Ghanem, S., El Shafey, H.M., Manzoor, N., 2018. Assessment of antimicrobial resistance patterns in Escherichia coli isolated from clinical samples in Madinah, Saudi Arabia. African Journal of Microbiology Research 12, 321-326.
CrossRef - Schechter, L.M., Creely, D.P., Garner, C.D., Shortridge, D., Nguyen, H., Chen, L., Hanson, B.M., Sodergren, E., Weinstock, G.M., Dunne, W.M., 2018. Extensive gene amplification as a mechanism for piperacillin-tazobactam resistance in Escherichia coli. Mbio 9, e00583-00518.
CrossRef - Shahid, M., Sobia, F., Singh, A., Malik, A., Khan, H., Jonas, D., Hawkey, P., 2009. Beta-lactams and beta-lactamase-inhibitors in current-or potential-clinical practice: a comprehensive update. Critical Reviews in Microbiology 35, 81-108.
CrossRef - Shobrak, M.Y., Abo-Amer, A.E., 2014. Role of wild birds as carriers of multi-drug resistant Escherichia coli and Escherichia vulneris. Brazilian Journal of Microbiology 45, 1199-1209.
CrossRef - Somily, A.M., Habib, H.A., Absar, M.M., Arshad, M.Z., Manneh, K., Al Subaie, S.S., Al Hedaithy, M.A., Sayyed, S.B., Shakoor, Z., Murray, T.S., 2014. ESBL-producing Escherichia coli and Klebsiella pneumoniae at a tertiary care hospital in Saudi Arabia. The Journal of Infection in Developing Countries 8, 1129-1136.
CrossRef - Springer, B., Kidan, Y.G., Prammananan, T., Ellrott, K., Böttger, E.C., Sander, P., 2001. Mechanisms of streptomycin resistance: selection of mutations in the 16S rRNA gene conferring resistance. Antimicrobial Agents and Chemotherapy 45, 2877-2884.
CrossRef - Tenaillon, O., Skurnik, D., Picard, B., Denamur, E., 2010. The population genetics of commensal Escherichia coli. Nature Reviews Microbiology 8, 207.
CrossRef - Willshaw, G., Cheasty, T., Rowe, B., Smith, H., Faithfull-Davies, D., Brooks, T., 1995. Isolation of enterotoxigenic Escherichia coli from British troops in Saudi Arabia. Epidemiology & Infection 115, 455-463.
CrossRef - Yoshizawa, S., Fourmy, D., Puglisi, J.D., 1998. Structural origins of gentamicin antibiotic action. The EMBO Journal 17, 6437-6448.
CrossRef - Zhang, X.Y., Ding, L.J., Fan, M.Z., 2009. Resistance patterns and detection of aac (3)-IV gene in apramycin-resistant Escherichia coli isolated from farm animals and farm workers in northeastern of China. Research in Veterinary Science 87, 449-454.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





