How to Cite | Publication History | PlumX Article Matrix
Rabha EL othmany , Hafida Zahir*
, Hafida Zahir* , Chorouk Zanane
, Chorouk Zanane , Doha Mazigh
, Doha Mazigh , Mostafa Ellouali
, Mostafa Ellouali and Hassan Latrache
and Hassan Latrache
Laboratory of bioprocess and biointerfaces, faculty of sciences and technics, sultan moulay slimane university, Morocco,23000.
Corresponding Author E-mail: hafidazahir@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2901
ABSTRACT:
Streptomyces has many advantages for exploration in biotechnological applications because of their ability to elaborate a multitude of bioactive molecules and secondary metabolites. Despite the importance of this genus in biotechnology, biofilm formation in Streptomyces is under-investigated. The objective of this research is to adapt two assays for the assessment of biofilm formation in Streptomyces. In the present investigation, we assess and follow biofilm formation in eight Streptomyces strains using quantitative and qualitative methods. The quantitative study based on a staining of the retained biomass in the microtiter plate with crystal violet “5%” and destaining using ethanol/acetone mixture, the concentration of crystal violet in the alcoholic solution reflect the intensity of the attached biofilm. On the other hand, the qualitative one consists of using modified freeman’s method a modified congo red agar method based on the color of colonies.
Quantification of biomass by crystal violet staining method confirmed that Streptomyces bellus A43 and Streptomyces bellus A61 are biofilm-forming and this ability increase with the period of incubation. Our results showed that sixStreptomyces strains arenon-slime producing/non-biofilm forming. Two Streptomyces strains are slime producing/biofilm forming; this character vanishes at five days. Further research on genes responsible for biofilm formation in Streptomyces is highly recommended for better understanding of the phenomenon.
KEYWORDS: Biofilm Formation; Congo red; Crystal violet; Slime; Streptomyces
Download this article as:| Copy the following to cite this article: Othmany R. E, Zahir H, Zanane C, Mazigh D, Ellouali M, Latrache H. Adaptation of Congo Red Agar Method and Microtiter Plate Assay to Study Biofilm Formation in Streptomyces. Biosci Biotech Res Asia 2021;18(1). |
| Copy the following to cite this URL: Othmany R. E, Zahir H, Zanane C, Mazigh D, Ellouali M, Latrache H. Adaptation of Congo Red Agar Method and Microtiter Plate Assay to Study Biofilm Formation in Streptomyces. Biosci Biotech Res Asia 2021;18(1).Available from: https://bit.ly/39Y1QfB |
Introduction
A Biofilm isdescribed as an irreversible association of microbes with biotic or abiotic surfaces1. The elaboration of the extracellular polymeric matrix (EPS), also called slime, is a determining factor in the detection of biofilms. The molecular composition of this EPS is specific to the microbial species2. According to Flemming, living inside biofilm community is part of the life cycle of most if not all bacteria. This lifestyle provides microbial cells-for a while- better survive in harsh environmental conditions3,4.In clinical settings, biofilms pose serious problems. They are commonly found on medical devices5,6also increases the resistance of microbial species responsible of nosocomiale infections7. In industrial fields, biofilms are responsible of clogging, blockages, energy transfer inefficiency. They are also responsible of surfaces corrosion5,6.
In wasterwater treatements, biofilm reactors are more efficient than others involving the use of sessile microorganisms, also in biotechnology/bioconversion field for the production of chemical substance with economic values 8. Curently, the manipulation of the biofilm in biorectors is difficult and poorly studied. However, they are used to respect economics and space-time yield 9,10.
Streptomyces, the representative genus in the phylum Actinobacteria, having the ability to produce valuable molecules and enzymes. Exploration of biofilm formation in this genus could provide crucial informations for controlling production of secondary metabolites and efficiency in environmental process.
The most commonly used methods for detecting and quantifying biofilms in bacteria are crystal violet staining methods (including microtiter plate and tube methods), congo red agar method, EPS assay, hydrophobic interaction chromatography assay, contact-angle measurements, bacterial adhesion to hydrocarbon assay, autoaggregation method and scanning electron microscopy, atomic force microscopy, confocal scanning microscopy…etc 11–16.
There is always an urgent need for easy and reliable methods as alternative to expensive technics. This research aims to test an easy and low-cost approach for screening of Streptomyces with ability to form biofilm for further purposes.
In the present study, two methods are tested with slight modification for screening biofilm formation ability. EightStreptomyces strains are tested. A modified congo red agar method for qualitative investigation, inspired from freeman’s method which is used for the first time for Streptomyces species and crystal violet assay for quantification of biofilm formed.
Materialand Methods
Growth conditions of Streptomyces strains
Streptomyces strains arebelonging to bioprocess and bio-interfaces laboratory are used in this study17. Strains arecultivated on Bennett liquid medium for further tests. Spore stock is obtained by cultivating Streptomyces strains on agar Bennett for 10 days.
Testing Biofilm formation ability
Microtiteration plates Assay
Biofilms were cultivated in microtiter plates following the modified method proposed by Stepanović18. In Brief, tests are carried out in micotiter plate 96-well flat bottom (polystyrene, orange scientific, France). Streptomyces strains are cultivated over-night in Bennett liquid media in erlemeyer flask at 28°C. Then optical density of the bacterial was measured at 600 nm wavelenght and the concentration adjusted to 0.02-0.03. Each well was filled in aseptic conditions with a volume of 200µl of cell suspension. Plates were incubated in 28°C. The wells has been filled after 24h and rinsed by 200 µl of sterile water three times in order to eliminate non adhered bacteria. After washing, the remaining adhered biomass was heat-fixed through exposition to hot air at 80°C for 30 min. Biofilm formation was evaluated at 1day, 2days, 3days, 4 days, 5days, 6 days and 7days. Wells filled only with liquid Bennett medium without bacterial cells are the negative control.
The attached biomass has been stained for 5 min by 200 µl per well of cristal violet (5%). Stain surplus was evacuated under tap water18. Then plates were dried, the cristal violet bound to the biomass was resolubilized using 200 µl per well of (ethanol/ acetone) solution (80/20) (v/v). The OD of the obtained solutions were measured at 595 nm using a microtiter plates reader. The OD595 values reflects the amount of attached biomass to the microtiter plate wells. Experiments were replicated three times.
The OD ranges were evaluated for all strains and controls. The cut off values (ODc) was calculated. It is characterized as the sum OD control and three standard deviations (SD).
ODcut off = range OD of negative control + (3SD of negative control).
For each microtiter plate, ODcut off value was calculated inedependentely.
Results are interpretated and strains were divided to four categories:
OD ≤ ODcut off indicates Non biofilm forming (0).
ODcut off <OD ≤ 2xODcut off indicates Weak biofilm producer (* or 1).
2xODcutoff<OD ≤ 4xODcut off indicates Moderate biofilm producer (** or 2).
4xODcut off <OD indicates Strong biofilm producer (*** or 3).
Adaptedcongo red method
Two phenotypic methods were tested to study biofilm formation by Streptomycesstrains. The qualitative study based on Congo red agar method,while the a quantitative oneused in this investigation is microtiter plate assay.
This test was performed following Freeman’s method with moderate adjustment16. The growth medium contain 10g/l of D-Glucose, 2g/l yeast extract, 1g/l meat extract, 2g/l peptone, 50 g/l sucrose, 10 g/l bacteriological agar and 1ml of autaclaved congo red (0.8g/l). Plates were inoculated and incubated at 37 °C.Then, the color of colonies was observed at 1 day, 2 days, 3 days, 4days, 5 days, 6 days, 7 days, 8 days, 9 days, and 10 days.19. Interactions of congo red with metabolites form complex product wich give darkness to colonies. Black colonies are categorized as biofilm-producers, whereas red colonies are considered as non-biofilm-formingbacteria. Tests are repeated 3 times.
Results and Discussion
Biofilm formation by streptomyces in microtiter-plates devices:
Figure 1 shows the quantification of biofilm formation for 7 days. Biofilm strength is reflected by the average of crystal violet optical density. As described in the methods part biomass attached to microtiter plate wells is stained using crystal violet. After dissolution using ethanol/acetone mixture crystal violet contained in the cells is found then in the alcoholic mixture. The optical density of this mixture measured 595nm reflect the amount of biomass adhered to well walls. Based on Stepanović classification 18 which refers to a comparison between Doc and DOtest it is possible to describe biofilm formation as strong, moderate, weak, or none.
 |
Figure 1: Quantification and following of biofilm formation Streptomyces strainsusing microtiter plate method. |
Streptomycesstrains (A3, A4, A10, A14, A15 and A23) areclassified as weak biofilm-forming strain, even after 7 days of incubation. Biofilms of those Streptomyces remain weak throughout the follow-up. On the other hand, Streptomyces bellus A43 and Streptomyces bellus A61 can form strong biofilm from 3 days of incubation. The evolution of the intensity of formed biofilms changes from weak to strong at 7 days. Streptomyces bellus A43 and Streptomyces bellus A61 are able to establish a strong attachment to wells. Colonization and kinetics of biofilm formation mainly depend on surface properties of both of Streptomyces and microtiter plate walls.
The measured density of the crystal violet reflects, in reality, the attached biomass to the wall which is composed of cells and extracellular matrix. Crystal violet is found in the microtiter plate wells after dissolution by ethanol/acetone. The high optical density of the mixture indicates a strong amount of adhered cells to the wells. According to the literature, few studies are conducted on the Streptomyces biofilm. In 2014, Winn characterized biofilm formation in the Streptomyces griseus ATCC 13273 cultivated in a tubular reactor; their results does not reveal any fixation of this strain in wells of polystyrene microtiter plates, they showed that the growth of this strain occurs at the liquid-air interface 20. Streptomyces biofilm formed in tubular reactor undergoes detachment at 24 h. According to Winn and collaborators, biofilm formation is a cyclical phenomenon; each attachment is followed by a detachment. These results 20 does not comply with ours which shows an increase of biofilm intensity by time incubation 20. However, Streptomyces was able to form stable biofilms during co-culture with Bacillus biofilm-forming 20 and Lactobacillus 21. This mixed culture lead to the formation of stable biofilms in bioreactors.
Ability of Streptomyces griseorubens and Streptomyces bellus strains to produce extracellular matrix (Slime).
Our tests are based on Freeman’s method CRA established on the differenceof colony color to distinguish between slime negative andslime positive bacteria (freeeman et al., 1989). The obtained results of the congo red test are presented in Table (1); as indicated the strains Streptomyces griseorubens A3, Streptomyces bellus A7, Streptomyces bellus A10, Streptomyces griseorubens A14, Streptomyces griseorubens A15 and Streptomyces griseorubens A23 showed a red colonies at 2 days of incubation (48h) which indicates the absence of slime production. While Streptomyces bellus A43 and Streptomyces bellus A61 reveal black colonies denoted consequently the presence of slime production.
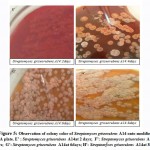
Following colony color evolution for 10 days leads to observe changes in slime production over time. Red colonies which appear in slime negative strains undergo a progressive modification, it varies from red to grey (Streptomyces griseorubens A3, Streptomyces bellus A7 and Streptomyces bellus A10 see Figure 2,3 and 4 ) or from red to white Streptomyces griseorubens A14and Streptomyces griseorubens A23(Figure 5 and 7).}
As known the genus Streptomyces has a complex developmental cycle including physiological and structural modification, spores germinate and give rise to vegetative hypha. The later is characterized by theability to anchor in the growth medium.A typical variation in cell surface properties occurs during the differenciation from vegetative growth to aerial growth. Aerial hypha risen during the aerial phase had typic color, it depends on the strain22–24 The observed color innon slime forming strainsand their derivates are due to the interactions of congo red with aerial hypha. Furthermore, these phenotypes observed in our case does not exist in Freeman’s or Arciola’s statements.
We note that Streptomyces bellus A43 and Streptomyces bellus A61 (Table 1, figure 8 and figure 9) at 2 days are considered as slime positive. Following the incubation, at 10 days it is showed that the black color changes to brown at 5days and became red at 9 days. It is suggested that slime formed at 2 days is used by those strains as a source of nutrients. The polysaccharides which interact with the congo red and give the dark color are probably metabolized.
Table 1: Phenotypic characterization and follow of slime production in Streptomyces griseorubens and Streptomyces bellus strains by modified congo red agar plate during 10days.
| 1day | 2 day | 3 day | 4 day | 5 day | 6 day | 7 day | 8 day | 9 day | 10 day | |
| A3 | No growth | Red | red | red | pink | white | white | grey | grey | grey |
| A7 | No growth | Red | red | pink | pink | white | white | grey | grey | grey |
| A10 | No growth | Red | red | red | white | white | white | white | white | grey |
| A14 | No growth | Red | red | red | white | white | white | white | white | white |
| A15 | No growth | Red | red | red | pink | pink | pink | pink | grey | grey |
| A23 | No growth | Red | red | red | pink | pink | pink | pink | grey | grey |
| A43 | No growth | Black | black | black | brown | brown | brown | brown | red | red |
| A61 | No growth | Black | black | brown | brown | brown | brown | red | red | red |
To our knowledge, there is no qualitative study elaborated based on congo red dye to describe biofilm formation in Streptomyces. However, several studies used congo red agar method to detect slime production for Staphylococcusstrains 2,25,26. The TSB broth supplemented with sucrose was most accurate medium for slime detection27. Mateo and collaborators investigated slime production in Staphylococcus epidermidis using congo red agar method 28. Although, the multitude of studies in slime detection, the mechanism by which interact congo red with EPS matrix and give black coloration still unknown16,29
The present study shows the effect of time incubation on slime production.No studies have been carried out on this parameter in Streptomyces or other bacteria. Following the incubation, at 10 days extracellular polysaccharides are probably metabolized. Our results demonstrate that slime production can be altered during incubationperiod; time is an important factor in the description of slime production. It is also proved that color of aerial hypha influence the interpretation since color indicated by freeman don’t includes those obtained in our case.
The complex developmental cycle of Streptomyces start with spore germination then development of aerial hypha and spores are formed and start a new lifecycle. Cell changes on cell Streptomyces surfaces are characterized by Atomic force microscopy, it was demonstrated that vegetative cell produce extracellular matrix, while aerial hypha are characterized by the absence of extracellular matrix 30. Treatments of Streptomyces pellets by DNase and protease demonstrate that extracellular matrix is mainly proteins20. The later suggestion is in agreement with Petrus investigation which approved the presence of rodlet layer composed mainly of proteins assembly31. The extracellular layer exist during aerial growth, its major role is to mediate the attachment to hydrophobic surfaces.
Many modified methods are proposed to detect biofilm formation through slime production. This modification occurs in medium composition 2,32. It is shown that accuracy is dependent on the growth medium. In Staphylococcus, it was found that accuracy is generally confirmed by analysis of intercellular aggregation locus (icaAB genes) 33.
Conclusion
To study biofilms in Streptomyces, slight modifications are performed to the congo red method and microtiter plate assay.We concluded that, the modified congo red agar method could be used to follow extracellular matrix production in Streptomyces just for 3 days, because the lifecycle of Streptomyces is complex and the color of aerial hypha interfer with the phenotypic characters. The aerial mycelia reveal a color charactristic to the Streptomyces strain. Theses colors that appears during aerial growth does not exist in the average proposed by Arciola or Freeman. Streptomyces bellus A43 and Streptomyces bellus A61are able to form strong biofilms from 3 days and the biofilm intensity increases over time. Streptomyces griseorubens A3, Streptomyces bellus A7, Streptomyces bellus A10, Streptomyces griseorubens A14, Streptomyces griseorubens A15 and Streptomyces griseorubens A23cannot form stable biofilm even after 7 days of incubation. The monitoring of biofilm formation by microtiter plate assayindicate that biofilm forming strains slime positive strains have the ability to establish strong biofilm at 3 days. The results obtained by congo red agar method are confirmed by microtiter plate assay. The combination of these easy methods could be an alternative to the complicated technics.
Acknowledgement
The authors recognize the help of the national center of scientific and technical research Rabat Morocco which provided us a scholarship.
Conflict of interest
The authors have no conflict of interest.
Funding Source
No financial support has been provided for this work.
References
- O’Toole, G., Kaplan, H. B. & Kolter, R. Biofilm Formation as Microbial Development. Annu. Rev. Microbiol.54, 49–79 (2000).
CrossRef - Arciola, C. R. et al. Detection of slime production by means of an optimised Congo red agar plate test based on a colourimetric scale in Staphylococcus epidermidis clinical isolates genotyped for ica locus. Biomaterials23, 4233–4239 (2002).
CrossRef - Flemming, H. & Wingender, J. The biofilm matrix. Publ. Gr.8, 623–633 (2010).
CrossRef - Kokare, C. R., Chakraborty, S., Khopade, A. N. & Mahadik, K. R. Biofilm : Importance and applications. 8, 159–168 (2009).
- Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nature Reviews Microbiology2, 95–108 (2004).
CrossRef - Flemming, H. C. Biofouling in water systems – Cases, causes and countermeasures. Applied Microbiology and Biotechnology59, 629–640 (2002).
CrossRef - Muffler, K., Lakatos, M., Schlegel, C., Strieth, D. & Kuhne, S. Application of Biofilm Bioreactors in White Biotechnology. (2013). doi:10.1007/10
CrossRef - Qureshi, N., Annous, B. A., Ezeji, T. C., Karcher, P. & Maddox, I. S. Biofilm reactors for industrial bioconversion processes : employing potential of enhanced reaction rates. 21, 1–21 (2005).
- Conroy, J. & Couturier, M. Dissolution of minerals during hydrolysis of fish waste solids. Aquaculture298, 220–225 (2010).
CrossRef - Asri, M., Elabed, S., Ibnsouda Koraichi, S. & El Ghachtouli, N. Biofilm-Based Systems for Industrial Wastewater Treatment. in Handbook of Environmental Materials Management 1–21 (Springer International Publishing, 2018). doi:10.1007/978-3-319-58538-3_137-1
CrossRef - Taj, Y., Essa, F., Aziz, F. & Kazmi, S. U. Original Article Study on biofilm-forming properties of clinical isolates of Staphylococcus aureus. J Infect Dev Ctries5, :403-409 (2012).
CrossRef - Wright, C. J., Shah, M. K., Powell, L. C. & Armstrong, I. Application of AFM from microbial cell to biofilm. Scanning32, 134–149 (2010).
CrossRef - Faust, J., Ba, M., Follo, M. & Wolkewitz, M. Biofilm formation and composition on different implant materials in vivo. 101–109 (2010). doi:10.1002/jbm.b.31688
CrossRef - Priester, J. H. et al. Enhanced visualization of microbial biofilms by staining and environmental scanning electron microscopy. Microbiol. Methods68, 577–587 (2007).
CrossRef - Christensen, G. D., Simpson, W. A., Bisno, A. L. & Beachey, E. H. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Immun.37, 318–326 (1982).
CrossRef - Freeman, D. J., Falkiner, F. R. & Patrick, S. New method for detecting slime production by coagulase negative staphylococci. 872–874 (1989).
CrossRef - Zanane, C., Latrache, H., Elfazazi, K., Zahir, H. & Ellouali, M. Isolation of actinomycetes from different soils of Beni Amir Morocco. Mater. Environ. Sci9, 2994–3000 (2018).
- Stepanović, S., Vuković, D., Dakić, I., Savić, B. & Švabić-Vlahović, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. Microbiol. Methods40, 175–179 (2000).
CrossRef - Arciola, C. R. & Baldassarri, L. Presence of icaA and icaD Genes and Slime Production in a Collection of Staphylococcal Strains from Catheter-Associated Infections. 39, 2151–2156 (2001).
CrossRef - Winn, M., Casey, E., Habimana, O. & Murphy, C. D. Characteristics of Streptomyces griseus biofilms in continuous flow tubular reactors. FEMS Microbiol Lett.352, 157–164 (2014).
CrossRef - Demirci, A., Pometto, A. L. & Johnson, K. E. Evaluation of biofilm reactor solid support for mixed-culture lactic acid production. Microbiol. Biotechnol. (1993). doi:10.1007/BF00167135
CrossRef - Chater, K. F. Genetics of Differentiation in Streptomyces. Rev. Microbiol.47, 685–711 (1993).
CrossRef - Mendez, C., Brana, A. F., Manzanal, M. B. & Hardisson, C. Role of substrate mycelium in colony development in Streptomyces. J. Microbiol.31, 446–450 (1985).
CrossRef - Hopwood, D. A. Streptomyces in Nature and Medicine. Oxford university press1, (2015).
CrossRef - Kaiser, T. D. L. et al. Modification of the Congo red agar method to detect biofilm production by Staphylococcus epidermidis. Microbiol. Infect. Dis.75, 235–239 (2013).
CrossRef - Croes, S. et al. Staphylococcus aureus biofilm formation at the physiologic glucose concentration depends on the S. aureus lineage. BMC Microbiol.9, 1–9 (2009).
CrossRef - Lee, J., Bae, Y. & Lee, S. Development of Congo red broth method for the detection of biofilm-forming or slime-producing Staphylococcus sp. LWT – Food Sci. Technol. (2016). doi:10.1016/j.lwt.2016.03.023
CrossRef - Mateo, M., Maestre, J. R., Aguilar, L. & Giménez, M. J. Strong slime production is a marker of clinical significance in Staphylococcus epidermidis isolated from intravascular catheters. 311–314 (2008). doi:10.1007/s10096-007-0433-y
CrossRef - Dadawala, A. I. et al. Assessment of Escherichia coli isolates for in vitro biofilm production. World3, 364–366 (2010).
- Del Sol, R., Armstrong, I., Wright, C. & Dyson, P. Characterization of changes to the cell surface during the life cycle of Streptomyces coelicolor. Atomic force microscopy of living cells. Bacteriol.189, 2219–2225 (2007).
CrossRef - Petrus, M. L. C. & Claessen, D. Pivotal roles for Streptomyces cell surface polymers in morphological differentiation, attachment and mycelial architecture. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol.106, 127–139 (2014).
CrossRef - Mariana, N. S., Salman, S. A., Neela, V. & Zamberi, S. Evaluation of modified Congo red agar for detection of biofilm produced by clinical isolates of methicillin – resistance Staphylococcus aureus. 3, 330–338 (2009).
- Dias, T. et al. Modi fi cation of the Congo red agar method to detect bio fi lm production by Staphylococcus epidermidis ☆. 75, 235–239 (2013).
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.