How to Cite | Publication History | PlumX Article Matrix
Bacteriophage Structure, Classification, Assembly and Phage Therapy
Department of Microbiology, Acharya Prafulla Chandra College, New Barrackpore, Kolkata-131. India
Corresponding Author E-mail: nabanita@apccollege.ac.in
DOI : http://dx.doi.org/10.13005/bbra/2911
ABSTRACT:
Current emergence of multidrug resistance and limitations in the development of the new antibiotics has proposed the problem of treating bacterial infections more challenging. This scenario may lead to the fear of failure in treating the multidrug resistant (MDR) bacterial infections and fuelled the uses of bacteriophages as an alternative of the conventional antibiotics in the post antibiotic era.So it is very much essential to know about the details of phage life cycle, assembly of phage complete structure, configuration and function of phage associated proteins etc. Although phages have been discovered a century ego, detailed study about lytic phages are gaining more interest in global fight against MDR bacterial species. This review has highlighted the basic knowledge of bacteriophage with the past and present scenario of several clinical studies targeting the MDR bacterial species. On the other hand it also discussed about the other uses of phages except human clinical trials.
KEYWORDS: Assembly; Bacteriophage; Classification; Multidrug Resistance; Phage Therapy
Download this article as:| Copy the following to cite this article: Giri N. Bacteriophage Structure, Classification, Assembly and Phage Therapy. Biosci Biotech Res Asia 2021;18(2). |
| Copy the following to cite this URL: Giri N. Bacteriophage Structure, Classification, Assembly and Phage Therapy. Biosci Biotech Res Asia 2021;18(2). Available from: https://bit.ly/3jbyqA5 |
Introduction
The crisis in clinical care imposed by multi-drug resistant (MDR) bacterial species causes a major concern in healthcare. The situation is particularly acute for rising of MDR pathogens for which few antibiotics are using currently(Gibson et al. 2019). The mechanisms that are responsible for the evolution of bacteria include interconnected travel, over use of antibiotics, transduction, mutations and conjugations. These suggest that the bacteria are very much flexible to embrace the newly discovered antibiotics within a short period of time.
Fortunately, “Bacteriophage”a natural alternative of chemical antibiotics has achieved an era of interests to overcome this situation(Stearns 2020).The name bacteriophage was derived from ‘bacteria’ and the Greek term ‘Phagein’ which means “eat” came up with Felix d’Herelle (1917).Approximately 1031 bacteriophage particlesare present globally and 4-50 % of the produced bacterial populations are killed every day by bacteriophages (Wommack et al. 2009). The fact that unlike antibiotics, phages can destroy target bacteria with specificity and has a self-limiting infection in the presence of these targets.This is essential for the application of phages to inhibit specific bacterial growth.It can be a natural, nontoxic and active substitute for antibiotic therapy. Phages are multiply exponentially only in the presence of the target host without disturbing the host’s normal flora. In the view of the massive rise in antibiotic resistance in several clinically important bacterial species, uses of lytic phages are gaining more and more attentions as a therapeutic alternative against infectious diseases.The objective of this review is to explore the fundamental knowledge of phage biology with a brief discussion on the impact of uses on human health.
Biology of the Bacteriophage
Bacteriophages or phages are the natural viral entities that can infect target bacteria and they are supposed to be the most abundant organism on earth. They are classified on the basis of their morphology and the genetic materials. Most of the phages are tailed phage with dsDNA which belongs to the order of Caudovirales. Chromosome of the tailed phages packaged into procapsid by similar DNA translocase molecular motors but the strategy of DNA replication and resulted genome end are not alike(Casjens and Gilcrease 2009). Adsorption of the phages depends on the nature and structure peculiarities of receptor on the host cell surface. Based on +the life cycle, phages are mainly categorized into virulent and temperate phages. Virulent phages follow the lytic life cycle. But temperate phages initiate with the lysogenic cycle and occasionally turned into the lytic cycle under specific conditions. Holin and lysine are the two key proteins used by the lytic phages to destroy their host cells.
Bacteriophage Classifications
About 96% of the reported bacteriophages belong to the Caudovirales order, which includes three families: Myoviridae that have contractile tail, Podoviridae that have short tail and Siphoviridae have non-contractile long tail(Kaliniene et al. 2017). Filamentous, cubic, and polymorphic phages belong to the same order and are classified into 10 small families which comprise of about 3.6% of the existing bacteriophages. Table 1 shows examples of phage morphologies (Ackermann 2009).
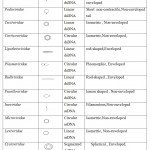 |
Table 1: Classification of Bacteriophages |
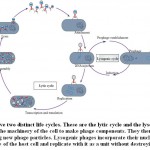
Phage Life Cycle
The first stage in infection is the adsorption of the phagesto the host cell surface receptor such as lipopolysaccharide (LPS), outer membrane proteins,flagella orfimbriae.This reversible adsorption process is facilitated by tail fibers or tail spikes of phages. Some of these fibers have enzymatic activity and are able to depolymerize the polysaccharides of the bacterial cell wall(Prokhorov et al. 2017). This reversible binding is followed by a base plate structural re-arrangement that triggers the contraction of tail-fibers. Binding becomes irreversible once the tail-fibers contract and bend toward the host cell to inject the virion genome into the cell cytoplasm. The ejection of the phage nucleic acid starts with the penetration of the tail tube into the host cell periplasm. During penetration, the virus uses a variety of lysins and carbohydrate de-polymerases. A transmem brane channel for phage DNA delivery is formed after the tail-fiber fuses with the host cytoplasmic membrane. Phage DNA is then ejected into host cytoplasm through this channel.
Bacteriophages possess several lifecycles; lysogenic or temperate, lytic or virulent, chronic andpseudolysogenic. In case of the lytic lifecycle, after infection,phage rapidly replicates and ends with the release of newly formed phage particles. Lytic infection effects in clear plaques on the host bacterial lawns. Temperate phages possess both lytic and lysogenic life cycle. In the lysogenic life cycle, phage DNA is incorporated into the bacterial chromosome by site-specific recombination or transposition which is called as prophage.In the integrated condition host cell can get new or additional properties upon expression of genes which is known as lysogenic conversion. For example, the cholera and shiga toxins are usually carried out and transported by prophage(Nanda et al. 2015).The prophages can be stable for many generations till environmental or physiological conditions trigger to the lytic lifecycle by induction process.As for example controlling of lysogeny and switching to lytic mode in λ phage is mediated by cl and cro genes(Court et al. 2007).Pseudolysogenyis the results of a successful adsorption and genome transfer eventof phages followed by non-replicative and non- productive statein adverse condition.
Chronic infection has been detected in some filamentous phages and archaeal viruses. In this case, progeny virions are slowly and continuously released from the cell surface rather than release as single burst phenomenon.
Lysogenic phages incorporate their nucleic acid into the chromosome of the host cell and replicate with it as a unit without destroying the cell.
Phage structure and assembly
Different bacteriophages have different sizes andshapes. Most well studied belong to the ds DNA containing phages. The tail phages have three major part; a capsid that filled with genome, a tail serve as a pipe for injection of genome to the host cell and a distinct adsorption device at the end of the tail which recognizes the host cell receptor. Assembly of dsDNA phages follow several steps: a. Assembly of prohead which is composed of capsid proteins filled with scaffolding proteins and portal proteins, b. Packaging of DNA, c. Maturation of proheads to mature heads, d. Connection of the neck and tail proteins.
Capsid organization
The mature infective phages have multiple factors including capsid, portal protein, scaffolds, terminase with other essential proteins. The capsids often have the structure of icosahedral symmetry(Tavares 2018). Different copies of capsid protein are usuallygathered into icosahedral shells. A minimum 60 copies of the capsid protein of identical environment is necessary to make and icosahedral particle. The number of the copies of the same protein within the independent part of the icosahedral lattice is represented by the triangulation number T. For example if T=3, virus has 180 subunits. Total number of viral capsid protein is described by T x 60.
Procapsid formation
The capsid where the DNAto be packaged is called procapsid or prohead.A capsid protein, with the help of scaffold protein forms the correct icosahedral geometry of capsid shell. Local concentration of scaffolding protein are primarily increased by the phage scaffolding protein that results into an entropic sink which promote association of coat proteins (Fane and Prevelige 2003).
In the phages,multi protein connector complex replace the fivefold vertices of the capsid (HTI,head-to-tail interface) comprising of portal protein and head completion protein. Phage genome is kept inside the capsid under high pressure by the control of HTT which serve as valve for closing and opening the channel (Tavares 2018). Portal protein is also involved into the entry and release of DNA along with the attachment of the neck protein to the head. Monomers of the different portal proteins vary in sizes and characterized by four domains: crown, clip, stemand wing. DNA packaging is facilitated by the clip domain that helps binding to the terminase.This domain is connected to the wing region by a stem that includesbasicallythe outer loops and two α-helices.The wing domain acts as a spine of the wing. Head assembly may bestarted from the portal tip by polymerization of the capsid and scaffolding proteins.
Capsid maturation
The scaffolding domain is cleaved off as soon as the procapsid is assembled, and removed from the capsid to make space for the genome. In lambda phage scaffolding protein exits from the capsid with the interaction of DNA, as lambda phage protease is genetically inactive. Release of scaffolding protein and starting of DNA packaging initiate the head maturation. It involves a large structural transition of prohead which results into the more stable bigger thinner shell head. During maturation, prohead can dissociate into subunits which allow proofreading and connection of misassembled intermediates. Many phages incorporate minor and pilot proteins into the head in the course of the head assembly with low copy numbers which are not essential for structure formation but crucial for virion infectivity. In phage P22 there are three minor proteins which localize in the shaft above the connector(Lander et al. 2006) T7 phages have an intricate shaft structure (inner core) which forms the capsid and consists of a protein ring with twelve fold symmetry. The release of terminase after DNA packaging mainly depends on the difference in the core structure.
Capsid assembly completion
After package of the DNA into the head, the terminase complex is exchanged by neck proteinand forms a gatekeeper complex together with connector which prevents the leakage of the mature DNA from the head and induces the exit of DNA upon attachment to the host. There are two types of neck proteins in Siphoviridae and Myoviridae phages,each make a ring below the portal protein. In SPP1 and HK97 like phages have the same ring closet structure to the connector but different fold presents in case of phage lambda whereas the second ring have the similar fold in SPP1 and lambda(Cardarelli et al. 2010).
Phage DNA packaging
Chromosome of tailed phages packaged into a procapsid by the way of DNA translocation but the strategy of DNA replication and the nature of packaged DNA ends are not same. Different types of characterized termini are (i) direct repeats with circular permutationat the terminal (ii)cohesive single-stranded ends (iii) direct terminal repeats with exact (non-permuted) long, several thousand base pairs, (iv)direct terminal repeats with exact (non-permuted) short, several hundred base pairs, (v) host DNA sequences at the terminal end, and (vi) terminal proteins bound covalently. Circular or replicating concatamer generates five of the above six types of virion chromosome ends by nucleolytic cleavage.DNA molecules replicate as monomeric linear molecules only in the case of phage genomes with terminal proteins. Phage-encoded enzyme called “terminase”perform these cleavages that are tightly coupled in all cases to the DNA packaging process. Those packaging mechanisms are followed by two steps-In first event terminase cleaves at pac site (Heedful packaging phage) or cos site (cohesive end phages) and the resulted DNA ends threaded into viral procapsid unidirectinally. There after a second neucleolytic cut (headful cleavage) occurred when DNA has filled the procapsid which results in the release of the packaged DNA from concatamer. Then second packaging event take place on that concatamer by inserting the unpackaged concatamer end in to a new procapsid. As like the first one the second event is terminated.Such unidirectional event is usually two five packaging event long. As per the packaging event the DNA of the tailed phages should be linear ds DNA because the threaded of the DNA into virion and release out from the virion is mediated by a very narrow portal passage which may not allow two parallel ds DNA simultaneously (for circular chromosome)(Casjens and Gilcrease 2009). Understanding the actual nature of the linear viral DNA,further studies are required.
Cohesive end
Phage λ,HK 97 are the well-studied COS phages. Both ends of the cohesive termini contain 7-19 nucleotides long protruding single strand phage chromosome which are complementary to each other by sequence. DNA ligaseanneals the ends to each other and each strand is closed to form covalently – closed circular molecules which serve as template for DNA replication after injection.Sequence specific, staggered cuts in the two DNA strand on a concatamerare made by terminase to generate the right end of the one chromosome and left of the subsequent chromosome to be packaged. Thus all the individuals of the COS phage having cohesive end termini are present on the genome sequenceat identical location (Casjens and Gilcrease 2009).
Head full packaging
P1,P22,SPP1, T1and T4phages contain circularly permutated and terminally redundant chromosomes which called “Headful packaging” phage. For phage P22, a sequence specific cleavage is took place by terminase at pac site on the replicated concatamer to initiate packaging series when procapsid is filled by DNA(Weigele et al. 2005). Upon infection the direct terminal repeats of the end goes for homologous recombination to generate the circular genome template for DNA replication. Cohesive ends are not present atheadful packaging phages because it is assumed that,as they are able to ligate to the other blunt ended DNA, the DNA ends are usually blunt.
Short exact repeats
Phages like T3 and T7 have direct double stranded repeats of several hundred base pair long same nucleotide at the termini of every virion chromosome (non-permutated). Duplication of the direct repeat DNA in concert with packaging produces the terminal repeats.
Long exact repeats
The phage like T5 and SP01 has several hundred base pair precise direct terminal repeats at the ends (Wang et al. 2005).How these long repeats are generated,the exact mechanism though is not yet cleared fully but phages are appear to be only one copy of the terminal between genomes in replication generated concatamerswith the exact short repeats, which suggest that the during packaging repeat region is duplicated.The best way to estimatethe position of the ends of such molecule on the sequence,restriction mapping are the best way to locate.
Host DNA at ends
Phage Mu replicates their genomes into host DNAby duplicativetransposition, which results into the packaging of viral genome that is integrated into host DNArandomly. Terminase recognizes a pac site which is located near one end of the genome and DNA Packaging event is initiated by cutting the nearby host DNA(Groenen and van de Putte 1985).
Covalently bound terminal protein
Phages like φ29 have covalently bound protein at the end of their virion and replicative chromosome. Terminal fragments of these type of DNA shows abnormal slow migration in electrophoresis gel while protease treatment restore the normal movement (Salas et al. 1978).
Tail assembly
The organization of the tail among the phages is different like; Podoviridae have very short tail, Siphoviridae have long non-contractile tailand Myoviridae have rigid long contractile tail. After the completion of the head assembly, the tail proteins are sequentially attached to the capsid. In Siphoviridae and Myoviridae phages, the tail is linked to the head via neck protein where in case of T4 phages attachment of the tail is followed by the preassembled fiber (Rossmann et al. 2004). Head and tail assembly occurred independently in a strict order of association of protein. There are different mechanisms which control that type of sequential attachment. Conformational switching is one of the most common mechanisms among them. The viral capsid with the T number higher than one showed the conformational switching when several quasivalent conformations are taken by the capsid protein. The initiator complex starts the assembly of tail in Siphoviridae and Myoviridae phagesthat forms the absorption at the distal end of the phage. six to eight proteins usually forms the long initiator complex in Siphoviridae and Myoviridae respectively. Base plate initiates the polymerization of the cylindrical section of the tail during the tail assembly,. The cylinder of the Siphoviridae is composed of multiple tail tube components. Lambda tail tube protein gpV cannot form a tubular structure without the initiator complex and exist as a monomer. Thetail tube protein of Myoviridae phage does not assembled into tubes as like lambda gpV without the base plate.
The tail assembly is completed after polymerization of the cylindrical part of the tail by the binding of the terminator proteins. The neck protein then interacts with the terminator proteins and induced the association of head to the tail. Functional and structure comparison of neck concludes that tail completion proteins and tail tube develop from a single ancestral gene. Tail tape measure proteins define the length of the tail.
Assembly completion
Assembly process of Siphoviridae and Myoviridaeare completed by the spontaneous joining of head and tail whereas binding of gpwacto the neck region forming whiskers prior to head to tail associationin case of T4. For the attachment of the preassembled tail fibers along with tail at the proximal part of the base plateswhiskers are very much necessary.In most of theSiphoviridae phages distal tail protein (DTP) form an oligomeric ring which is attached to the tail tube(Veesler and Cambillau 2011). Recognition and attachment to receptor binding proteins are served by DTP, sometimes in T5, T4 and other phages tail fibres assisted this interaction. Some of the Gram-negative bacteria specific phages have lysozyme at the end of their tails and digest the peptidoglycan barrier after entry into the periplasm.
Cell Lysis
The last event of the infection cycle is thelysis of the bacterial cells of a lytic phage. The time between the infection and production of the progeny is called the latent period. Optimal time of the lysis dependsupon the quality (physiological state) and quantity (density) of the host bacteria.The optimum latent period is shorter under the highest host quality or quantity. Phages have to overcome several host barriers to release the accumulatedvirion particles in the environment, later which infect the other available host. The cytoplasmic membrane, cell wall and in case of Gram negative bacteria, the outer membrane are the barriers. Phages have evolved several strategies to overcome those barriers.Phages (dsDNA) encodeholin protein which forms lesion in the cell membrane. Another phage coded protein endolysin (muralyticenzymes) is able to attack the cell wallas a result of the disruption of the cell membrane which results in the lysis of the host cell for the release of the virion particles (Catalao et al. 2013). Among the well-studied lytic phagesthe holin-endolysin system is very common but not found in ssDNA or ssRNA phages.
Identification of the Host Cell Surface Receptor
Identification of host cell surface receptor is the crucial for the adsorption which confers the specificity of the host among the phages. Such specificity is dependent on the nature and structural particularities of receptors on bacterial cell surface(Rakhuba et al. 2010). Host cell surface of Gram negative bacteria is characterized by complex lipopolysaccharide (LPS) and protein structure. Out of them either LPS or protein served as a receptor. In some cases LPS and protein part both may serve as receptor.
Phage Therapy
Emergence of antimicrobial resistance and limitations in development of new antibiotics is the biggest threat to global health.WHO in 2019 stated that the increasing rate of MDR bacterial species requires serious actions all over government sector and society(Organization 2019). Estimated MDR bacterial species could kill more than 10 million people per year. So in this situation finding out of an alternative to antibiotics is too much essential.Recently Govt. of India has launched a programme “Antimicrobial Resistance Hub” in 2019 to combat those MDR pathogens which give priority on the use of phages as a therapeutic agent. Bacteriophages are the natural predator of the bacteria found ubiquitously in nature which are non-toxic and host specific. The multidrug resistant (MDR) bacterial species that cause clinically relevant bacterial infections can be destroyed by the administration of the bacteriophages. This is called phage therapy.
Although early report of phage therapy,the introduction of antibiotics affected primarily but in the post antibiotic era,well-deserved rebirth is emerging. Some organizations which are devoted to phage therapy areMicrobiology and virology (Georgia),Eliava phage therapy centre,Gangagen (India), Institute of immunology and experimental therapy (Poland), Intralytix (USA) and so on.
Early Phage Therapy
Bacteriophages from human faces was first isolated and used them to kill bacteria including Shigella spp.by Felix d’Herelle and his co-workers in 1917 under laboratory condition and they developed the idea of ‘‘phage therapy’’ (d‘Herelle, 1917). In 1921 patients suffering from dysentery at Paris were cured by oral phage applications wit in one day. In Alexanderia patients suffering from bubonic plague were allowed for phage treatment by injection of phages directly to the lymph nodes, the site of infection. In 1927 during cholera epidemic in India the mortality rate of the phage treated patients was reduced to 8% from 68%. Phage therapy was massively implanted on the Soviet soldiers during the winter war between Soviet Union and Russia (1939-1940) against open wound with Staphylococcal or Streptococcal infection which prevent amputation. Staphylococcal or Streptococcal phages with or without variety of phages including Salmonella phage, Shigella phage, E.coli phage, P.aeruginosaphage were used to treat the wounds which results in the reduction of death rate by Gangrene. Thereare several organizationswhich currently use phages as therapeutic agent (Table 2).
Table 2: Key Features and Deeds of Phage Therapy Centers
| Name of center | Country | Main features and deeds |
| Center for Phage Therapy | Poland | 1,500 patients with supportive bacterial infections have been cured by specific bacteriophages, from 1980, routine antibiotic therapy ineveryplace has unsuccessful (http://www.iitd.pan.wroc.pl). |
| Eliava Phage Therapy Centre | Georgia | bacteriophage preparations have been established by agroup of eight laboratories for fighting against perilous and antibiotic-resistant superbugs
(http://www.phagetherapy.com). |
| Novomed | Georgia | Not only the local Georgians avail the treatments, but also to foreign patients with chronic wounds and other types of acute infections.
(http://www.mrsaphages.com). |
| Phage Therapy Centre | Georgia | Provides exceptional treatment for patients having bacterial infections and chronic, drug-resistant or not responded to conventional antibiotic therapies (http://www.phagetherapycenter.com). |
Current Phage Therapy
The effects of phage therapy are continuously publishing by the Russia, Poland and Eliava institutes and that helped to keep this approaches popular.The unanswered question of phge therapy is now gradually addressed resulting in the phage therapy gaining more popular and interesting. Phages have been administrated topically,orally (tablets or liquid), intravenously and rectally for more than 90 years in Eastern Europe without serious side effects. Phage therapy are now offered by several clinics in different countries to treat diseases(Table3 and 4)(Duplessis CA et al. 2019; Kortright et al. 2019).
Table 3: Case Study of Phage Therapy
| Sr.No | Infection | Complicating Conditions | Antibiotic Resistance | Phage Dose and Application | Duration of Phage Treatment | Outcome |
| 1 | Pseudomonas aeruginosabacteremia | DiGeorge syndrome, congenital heart disease with pacemaker | Meropenem,tobramyci, aztreonam, polymyxin B, colistin, Cephalosporins, and fluoroquinolones | 3.5 x105 PFU delivered intravenously every 6 h. | Initial treatment for 36 hours (six doses total), treatment resumed 11 days later | Blood cultures negative after phage treatment; returned to positive following termination of phage administration |
| 2 | Acinetobacterbaumanii infected pseudocyst (3 months) | Necrotizing pancreatitis | Cephalosporins,
meropenem, gentamicin, amikacin, trimethoprimsulfamethoxazole, tetracycline, ciprofloxacin, and colistin |
5 x109 PFU delivered intravenously every 6 h | 84 days (336 doses total), minocycline added on day 2 | Clinicalimprovement of infection |
| 3 | Pseudomonas aeruginosa infected aortic graft (3 years) | Aorto-cutaneous fistula | Ciprofloxacin | 1 X 108 PFU topically delivered on fistula | Single dose |
four weeks post treatment Cultures negative; no relapse of infection after>2 years |
| 4 | Burkholderiacenocepacia | Cystic fibrosis s/p bilateral lung transplant Post-transplant pneumonia and sepsis | Ceftazidime,Amoxicillin,Tetracycline,Cethromycin,Sulfomethoxazole | One phage treatment comprising “1” Phage | 2 doses | No effect (Only two doses administered prior to expiring) |
| 5 | Pseudomonas aeruginosa | Left ventricular assist device infection and septicaemia | Cephalosporin,Penicillin,Carbapene,Polymixin B,Tobramycin,Levofloxacin | One phage treatment comprising “3” phages | 6 weeks |
Negative Blood cultures but not durable |
| 6 | Klebsiellapneumoniae and Acinetobacter. baumannii | Post-surgical knee wound infection | Carbapenem, Cephalospori, Quinolones, Ciprofloxaci, TigecycillinePolymixin B | K.
pneumoniaeone phage treatment comprising 1 phage A. Baumanii two treatments each comprising 1 phage |
2 weeks | Negative blood cultures |
| 7 | Burkholderiadolosa | Cystic fibrosis s/p bilateral lung transplant | Tobramycin, trimethoprim, Ceftazidime, Amoxicillin, Sulfomethoxazole | One phage treatment comprising1 phage | 5 weeks | Negative blood and BAL cultures |
| 8 | ESBL E. coli | Bacteremia/ urosepsis s/p kidney transplant | Penicillin, amikacin | One phage treatment comprising 2phages | 2 weeks |
Negative blood and urine cultures |
Table 4: Clinical trial of Phage Therapy
| Trial | Infection | Treatment Group | Phage Dose and Application | Outcome |
| 1 | Pseudomonas aeruginosa otitis | 12 individuals had received phage cocktail | 109 PFU was intra-aurally delivered (single dose) | From each group three individuals found to have undetectable levels of P. aeruginosa at the finale of the trial |
| 2 | Escherichia coli diarrheal diseases | 40 individuals had received phage cocktail M, 39 individuals had received phage cocktail T | 1.4 X 109 PFU cocktail M or 3.6 X 108 PFU cocktail T delivered orally three times per day for 4 days (12 doses)in oral rehydration solution | No substantial difference observed between placebo group and phage treatment group |
| 3 | Pseudomonas aeruginosa burn wound infection | 12 individuals received a phage cocktail | 2 X 107 PFU (expected) 200– 2,000 PFU (actual) topically applied one time per day upto 7 days (seven doses) | Trial halted, insufficient efficacy; may be due to considerably lower applied dose of phage than estimated |
| 4
|
Several multidrug-resistant bacteria | 157 patients was orally, intrarectally or intravaginally administrated to Hirszfeld Institute phage collection | 10- 20 ml of phage collection were administrated thrice daily for 12 weeks. | No antagonistic events; phage therapyprovided good response up to 40% rate |
| 5
|
Escherichia coli; Staphylococcus aureus; Pseudomonas aeruginosa | Intralytix phage cocktail WPP-201 were applied on 65 patients | 1 X 109 Phage cocktail were topically applied on wound infection once a week for 12 weeks | Different wound healing rates with time (differential wound size reduction over time) |
Other Uses of Phages
Apart from the alternative of antibiotics bacteriophages are also being used in different sectors such as livestock and food industries (Polaska and Sokolowska 2019).
Livestock
Salmonella: Acocktail of four phages were used in a boiler chick model to reduce the load of the Salmonella spp. which results in the reduction of 1log10 colony forming unit (CFU) after 14 days of administration.Saezet .al reported that administration of phages reduce the colonization andshedding of Salmonella in pigs(Saez et al. 2011).
Colibacilosis: Spray of two high titer phages reduce the colibacilli associated mortality in poultry(Huff et al. 2002).
Campylobacteriosis:Wagenoar et.al reports that administration of a phage preparation by oral gavage could protect 10 days old Ros broiler chick and adult chickens from campylobacteriosis caused by Campylobacter jejunii. In another report C.jejuni count in the ceca and upper intestine of phage treated birds were reduce between 0.5 and 5 log10 CFU/g(Loc Carrillo et al. 2005).
Clostridiosis: In a study with 900 birds showed that cocktail of five phages (105 PFU/ml) administration by oral gavage or drinking water against Clostridium perfringensinfection resulted in a 92 % mortality reduction than untreated control group(Miller et al. 2010).
Conclusion
Recently the emergence of multidrug resistant bacteria is a worldwide problem. Limitations in the development of new antibiotics made the problem more challenging. In this scenario bacteriophages are the promising therapeutic tools alternative to the antibiotics.For that, knowledge about the bacteriophage structure, assembly, phage packaging and phage genetics are very much essential. In this review, this particular aspect has been described in detail. Genetically modified or cocktail/combination of phages are being used to treat the variety of bacterial infections that are resistant to the latest used antibiotics. Several case studies have been carried out where microbiological eradication of the targeted organisms found after the administration of phages. Pathogens of the intensively-reared livestock also have been reduced after the uses of phages. However we cannot conclude that phages are appropriate to clear all the infections and curing of the clinical syndrome. Therefore, more clinical trials with several studies are needed to address the several issues coupled with the phage therapy such as evolutionary strategy, mosaic architecture andinvolvement of any phage gene in host pathogenesis. Lastly, the promising result of the previous study indicates that phage therapy may be a future remedy against MDR bacterial species.
Acknowledgment
Authors thank to DST-SERB, Govt. of India and Department of Biotechnology West Bengal Govt. for financial assistance.
Conflicts of interest
Author declares that she have no conflict of interest
Funding Source
DST-SERB, Govt. of India and DBT, Govt. of West Bengal.
References
- Gibson SB et al. (2019) Constructing and Characterizing Bacteriophage Libraries for Phage Therapy of Human Infections Frontiers in microbiology 10:2537 doi:10.3389/fmicb.2019.02537
CrossRef - Stearns SC (2020) Frontiers in Molecular Evolutionary Medicine Journal of molecular evolution 88:3-11 doi:10.1007/s00239-019-09893-5
CrossRef - Wommack KE, Bench SR, Bhavsar J, Mead D, Hanson T (2009) Isolation independent methods of characterizing phage communities 2: characterizing a metagenome Methods in molecular biology 502:279-289 doi:10.1007/978-1-60327-565-1_16
CrossRef - Casjens SR, Gilcrease EB (2009) Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions Methods in molecular biology 502:91-111 doi:10.1007/978-1-60327-565-1_7
CrossRef - Kaliniene L et al. (2017) Molecular Analysis of Arthrobacter Myovirus vB_ArtM-ArV1: We Blame It on the Tail Journal of virology 91 doi:10.1128/JVI.00023-17
CrossRef - Prokhorov NS et al. (2017) Function of bacteriophage G7C esterase tailspike in host cell adsorption Molecular microbiology 105:385-398 doi:10.1111/mmi.13710
CrossRef - Nanda AM, Thormann K, Frunzke J (2015) Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions Journal of bacteriology 197:410-419 doi:10.1128/JB.02230-14
CrossRef - Court DL, Oppenheim AB, Adhya SL (2007) A new look at bacteriophage lambda genetic networks Journal of bacteriology 189:298-304 doi:10.1128/JB.01215-06
CrossRef - Tavares P (2018) The Bacteriophage Head-to-Tail Interface Sub-cellular biochemistry 88:305-328 doi:10.1007/978-981-10-8456-0_14
CrossRef - Fane BA, Prevelige PE, Jr. (2003) Mechanism of scaffolding-assisted viral assembly Advances in protein chemistry 64:259-299 doi:10.1016/s0065-3233(03)01007-6
CrossRef - Lander GC et al. (2006) The structure of an infectious P22 virion shows the signal for headful DNA packaging Science 312:1791-1795 doi:10.1126/science.1127981
CrossRef - Cardarelli L et al. (2010) Phages have adapted the same protein fold to fulfill multiple functions in virion assembly Proceedings of the National Academy of Sciences of the United States of America 107:14384-14389 doi:10.1073/pnas.1005822107
CrossRef - Weigele PR, Sampson L, Winn-Stapley D, Casjens SR (2005) Molecular genetics of bacteriophage P22 scaffolding protein’s functional domains Journal of molecular biology 348:831-844 doi:10.1016/j.jmb.2005.03.004
CrossRef - Groenen MA, van de Putte P (1985) Mapping of a site for packaging of bacteriophage Mu DNA Virology 144:520-522 doi:10.1016/0042-6822(85)90292-2
CrossRef - Salas M, Mellado RP, Vinuela E (1978) Characterization of a protein covalently linked to the 5′ termini of the DNA of Bacillus subtilis phage phi29 Journal of molecular biology 119:269-291 doi:10.1016/0022-2836(78)90438-2
CrossRef - Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG (2004) The bacteriophage T4 DNA injection machine Current opinion in structural biology 14:171-180 doi:10.1016/j.sbi.2004.02.001
CrossRef - Veesler D, Cambillau C (2011) A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries Microbiology and molecular biology reviews : MMBR 75:423-433, first page of table of contents doi:10.1128/MMBR.00014-11
CrossRef - Catalao MJ, Gil F, Moniz-Pereira J, Sao-Jose C, Pimentel M (2013) Diversity in bacterial lysis systems: bacteriophages show the way FEMS microbiology reviews 37:554-571 doi:10.1111/1574-6976.12006
CrossRef - Rakhuba DV, Kolomiets EI, Dey ES, Novik GI (2010) Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell Polish journal of microbiology 59:145-155
CrossRef - Duplessis CA SM, Hamilton T MG, Brownstein M, et, al. (2019) A Case Series of Emergency,Investigational New Drug Applications for Bacteriophages Treating Recalcitrant,Multi-drug Resistant Bacterial Infections:Confirmed Safety and a Signal of Efficacy Intensive & Crit Care 5
- Kortright KE, Chan BK, Koff JL, Turner PE (2019) Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria Cell host & microbe 25:219-232 doi:10.1016/j.chom.2019.01.014
CrossRef - Polaska M, Sokolowska B (2019) Bacteriophages-a new hope or a huge problem in the food industry AIMS microbiology 5:324-346 doi:10.3934/microbiol.2019.4.324
CrossRef - Saez AC, Zhang J, Rostagno MH, Ebner PD (2011) Direct feeding of microencapsulated bacteriophages to reduce Salmonella colonization in pigs Foodborne pathogens and disease 8:1269-1274 doi:10.1089/fpd.2011.0905
CrossRef - Huff WE, Huff GR, Rath NC, Balog JM, Donoghue AM (2002) Prevention of Escherichia coli infection in broiler chickens with a bacteriophage aerosol spray Poultry science 81:1486-1491 doi:10.1093/ps/81.10.1486
CrossRef - Loc Carrillo C, Atterbury RJ, el-Shibiny A, Connerton PL, Dillon E, Scott A, Connerton IF (2005) Bacteriophage therapy to reduce Campylobacter jejuni colonization of broiler chickens Applied and environmental microbiology 71:6554-6563 doi:10.1128/AEM.71.11.6554-6563.2005
CrossRef - Miller RW, Skinner EJ, Sulakvelidze A, Mathis GF, Hofacre CL (2010) Bacteriophage therapy for control of necrotic enteritis of broiler chickens experimentally infected with Clostridium perfringens Avian diseases 54:33-40 doi:10.1637/8953-060509-Reg.1
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.