How to Cite | Publication History | PlumX Article Matrix
Jawed Shaikh1, Ashfaque Mehboob Khan1 and Mirza Mushtaq Vaseem Baig2*
1Department of Botany, Maulana Azad College of Art, Science and Commerce, Dr. Babasaheb Ambedkar Marathwada University, Aurangabad, India.
2Department of Botany and Department of Biotechnology, Yeshwant Mahavidyalaya, Nanded, India.
Corresponding Author E-mail: mmvbaig@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2913
ABSTRACT: Fungal endophytesare colonized in different part of the plants and play important role in survival of plants in stressful habitat. In search of potential endophytic fungito produce bioactive metabolites inthis study we investigate thediversity of endophytic fungi associated with leaves of the Acacia nilotica plant. Twenty-six endophytic fungi were subjected to morphologicaland molecular identification with internal transcribes spacer (ITS) region sequenced. All 26 endophytic fungi were divided into nine genera Chaetomium, Amesia, Ovatospora, Penicillium, Phialemonium, Colletotrichum, Crinipellis, Acrophialophora, Cribbea. Most of them belonged to the phylum Ascomycota only one belonging to the phylum Basidiomycota. This study shows that Acacia leaves inhabitant by diverse group of endophytic fungi. The biodiversity analysis showed Chaetomium sp. Being dominant with the highest colonization frequency (26.9%). One of the Chaetomium sp. showed sequence similarity of93% with the species reported earlier, Further investigationsarein needed to harness the bioactive compounds.
KEYWORDS:
Acacianilotica; Chaetomiumsp; Diversity; Endophytic fungi; IT Ssequencing; Leaf
Download this article as:| Copy the following to cite this article: Shaikh J, Khan A. M, Baig M. M. W. Diversity and Molecular Characterization of Endophytic fungi associated with Leaves of Acacianilotica. Biosci Biotech Res Asia 2021;18(2). |
| Copy the following to cite this URL: Shaikh J, Khan A. M, Baig M. M. W. Diversity and Molecular Characterization of Endophytic fungi associated with Leaves of Acacianilotica. Biosci Biotech Res Asia 2021;18(2). Available from: https://bit.ly/3y9e7YR |
Introduction
The microorganisms that colonize living plant tissues internally not inducing any overt symptoms or injury to the host plant are endophytes1. They are found in the diverse geographical region ranging from alpine plants to tropical plants including an ecologically adapted plants from hydrophytes to xerophytes. It is speculated that some of the tropical woody plants show hyper diversity of endophytic fungi2 .According to Surya narayana3, this diversity could be attributed to the host abundance or endophyte assemblage in a host. The diversity with host, environment, geography, and tissue type enables endophytes to produce a plethora of bioactive secondary metabolites4. In order to harness the bioactive compound from this cryptic resource, the study of the diversity of endophytes in different plant hosts from different regions is essential.
The relations of endophytes with plant hosts may be symbiotic to antagonistic5. These endophytes influence plants’ nutritional level, Survival, and distribution of plants at different stages6. Endophytes produce biologically active secondary metabolites7 such as VOCs(Volatile Organic Compounds)8, antidiabetic, anticancer, andanti-bacterial9, insecticidal active compound10. Endophytes can also produce structurally diverse secondary metabolites11. Aspergillus awamori, an endophytic fungus was characterized from the Acacia nilotica recorded for the synthesis of antidiabetic peptides12. The antimalarial bioactive metabolite epoxycytochalasin H against Plasmodium falciparum reported from endophytic fungus Diaporthe miriciae13. Two new antiviral compounds, cytonic acid A (C32H36O10) and B (C32H36O10) isolated from endophytic fungi Cytonaemasppalso reported14.
Most of the species of Chaetomium are members of non-clavicipitaceous endophytes that had gained the attention of investigator sowing tothe production of bioactive metabolites. Sharma etal.15 described a new species isolated from Jatropha podagrica in Indiaas C. jatrophae. Chaetomium iranianum and C. grande were isolated and characterized from Egypt and Africa by Blanchette etal.16 and Abdel-Azeem etal.17 respectively from the host Teucrium polium.
Acacia nilotica (L.) Del. belonging to the family Mimosaceae was selected to investigate the diversity of endophytes associated. The plant is distributed over a wide area in tropical and subtropical regions and found naturally growing throughout the Marathwada region. Leaf of A. nilotica shows pharmacological potential such as Chemopreventative, antimutagenic, antibacterial, anticancer18 astringent, anti-microbial, Aphrodisiac, dressing of ulcers 19 anti-inflammatory and treatment against Alzheimer’s diseases have been well documented 20.To study the induced secondary metabolites by these endophytes, it was thought proper to investigate the endophytes first, the aim of this study was to isolate and characterize fungal endophytes associated with leaves using both morphological methods as well as molecular methods based on genomic DNA’s ITS (Internal Transcribed Spacer) region.
Materials and methods
Isolation of fungal endophytes
Plant leaflets were collected and cleaned under running tap water to remove soil debris/ dust from the surface and then with ethanol (70%) for surface sterilization for 3 minutes, 1% sodium hypochlorite for 1 minute, followed by 3 rinses in sterile distilled water for 3 minutes each. Sterilized leaflets (0.5–1.0 cm) were plated on PDA Petri plates. The incubation of the plates was done at 28⁰C temperature for 3–4 weeks and were observed for emergence of colonies. The fungi from these colonies were subcultures to obtain pure isolates.
Morphological identification
Morphological Identification of the isolated endophytic fungi was done based on colony characters comprising of colour of colony, shape of colony, sizeof colony, and hyphal characters by using the identification key and microscopical observation of slides stained by cotton blue-stained slides.
Molecular characterization of endophyte
The nuclear DNA extraction was carried out using a spin column kit (Hi-Media, India). The amplification of Internal Transcribed Spacer rRNA gene (600 bp)21of the fungi was carried out using GeneAmp™ PCR System 9700. Exonuclease I – Shrimp Alkaline Phosphatase (Exo-SAP)22 treatment was used for further purification.The purified products of PCR were sequenced in ABI 3500xl genetic analyzer (Life Technologies, USA). The resulting Sequences were then analysed using the Basic Local Alignment Search Tool (BLAST), and the most related sequences were retrieved from GenBank NCBI23.The program compared nucleotide sequences to sequences from the relationshipdatabases and calculated the matches’ statistical significance, the result of which are given.
Phylogenetic tree
The relationship among the isolates was inferred by the Maximum Like lihood method and Tamura 3-parameter model24. The consensus tree was generated from 500 replicates 25 with bootstrapping from the tax a analyzed 25. The branches with less than 50% bootstrap replicates were merged. In the bootstrap test (500 replicates), the percentage of replication trees in which the related taxa clustered together is shown next to the branches 26. The Neighbor-Joining method was applied to a matrix of pairwise distances calculated using the Tamura 3 parameter model to produce the initial tree(s) for the heuristic search.There were 07 nucleotide sequences in this study. 1st+2nd+3rd+Non coding codon positions were included. In the end, the dataset contained 601 positions. MEGA X was used to run the evolutionary analyses.27
Statistical analysis
Hata and Futai’s formula was used to calculate the colonisation frequency (percent CF) of endophytic fungi23.
Percent Colonization Frequency(%CF)=(Ncol/Nt)100
Where,
Ncol=the number of plant tissue segments colonised by each fungus
Nt=the total number of plant tissue segments examined.
Results
Isolation of endophytic fungi
Endophytic fungi were isolated using potato dextrose agar (PDA) media. From 50 leaflets of A. nilotica, a total of 26 endophytic fungal species were isolated.
Morphological identification
The isolate AN2 exhibited morphologically as dense colony, with aerial habit and white coloured colony on PDA plate (Fig.1). Further, observation of the fungus through microscope showed the occurrence of hair-like, brown coloured appendages (setae) on their surface (Fig. 2). The morphological character of the isolated fungus was similar with the morphological structures of Chaetomium pachypodioides.AN17 isolate shows light brown and reverse yellowish-brown coloured (Fig 1) and microscopic observation shows the presence of hair-like, dark coloured appendages (setae) and lateral hairslike structures matching with the morphological features of Chaetomium Microthecia (Fig 2).
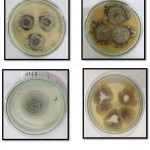 |
Figure 1: Endophytic fungi grow in PDA medium. |
 |
Figure 2: Microscopic images of endophytic fungi stained with Cotton blue stain.Images represent A. AN 2, B. AN 17, C. AN 11. |
Molecular characterization of endophytes
The ITS1–5.8S–ITS2 sequences of the most similar endophytic fungal isolates were retrieved from GenBank (NCBI), and phylogenetic tree (Fig 3) was created.The sequences showed different taxonomic affinities between the endophytic isolates extracted from GenBank. These sequences were submitted to GenBank, NCBI, with accession numbers listed in Table 1.
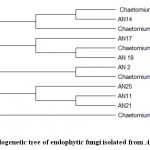 |
Figure 3: Phylogenetic tree of endophytic fungi isolated from Acacia nilotica. |
Table 1: NCBI- Accession number, %Sequence similarity and % Query Cover.
| Endophytic Isolates (GenBank Acc. no. of the ITS sequence) | Query coverage | % sequence similarity | Organism with the highest sequence identity, GenBank Acc. No. |
| AN2 (MW238764) | 99% | 96% | Chaetomium pachypodioides |
| AN11 (MW239168) | 100% | 93% | Chaetomium globisporum |
| AN14 (MW239165) | 100% | 100% | Chaetomium globosum |
| AN17 (MW242834) | 100% | 100% | Chaetomium microthecia |
| AN18 (MW242837) | 100% | 100% | Chaetomium pseudoglobosum |
| AN21 (MW248483) | 100% | 93% | Chaetomium globisporum |
| AN25 (MW242840) | 100% | 93% | Chaetomium globisporum |
Statistical analysis
The biodiversity analysis exhibited that the fungalcolonization by these endophytes was significant and diverse in leaf.Graphical representation is done by using R-studio software. Chaetomiumsp.showed the highest colonization (26.9%) frequency, (Table 2 and Fig 4).
Table 2: % Colonization frequency of endophytic fungi isolated from Acacia nilotica.
| Sr. No | Endophytic fungi | No.of Isolates | % CF |
| 1 | Chaetomium sp. | 07 | 26.9 |
| 2 | Amesia sp. | 05 | 19.2 |
| 3 | Ovatospora sp. | 03 | 11.5 |
| 4 | Penicillium sp. | 02 | 7.6 |
| 5 | Phialemonium sp. | 02 | 7.6 |
| 6 | Colletotrichum sp. | 01 | 3.8 |
| 7 | Crinipellis sp. | 01 | 3.8 |
| 8 | Acrophialophora sp. | 01 | 3.8 |
| 9 | Cribbea sp. | 01 | 3.8 |
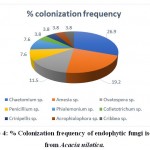 |
Figure 4: % Colonization frequency of endophytic fungi isolated from Acacia nilotica. |
Discussion
The alarming increase in microbial resistance has led the researchers to find out the novel sources of antimicrobial agents to control the drug resistant pathogens. Keeping this in mind, endophytic fungi were isolated from leaflet of the A. nilotica, followed by the proper surface sterilization. A total of 26 endophytic fungal species were isolated from 50 leaflets. Potato dextrose agar (PDA) media was used to isolate endophytic fungi. Out of total isolates of endophytic fungi (n=26) isolated from the host, most of them belonged to Phylum Ascomycota followed by one species of Crinipellis belonging to phylum Basidiomycota. Ascomycota phylum includes endophytic fungi from classes such as Sordariomycetes, Eurotimycetes, and Incertaesedis. The ITS1–5.8S–ITS2 sequences of all nearest neighbours of the endophytic fungal isolates were retrieved from GenBank (NCBI) and phylogenetic tree was constructed. The sequences studied showed diverse taxonomic affinities in the isolates. The same were submitted to NCBI and obtained accession numbers. Five isolates AN23 (Crinipellistabtim), AN11(Chaetomium globisporum), AN7 (Phialemoniumobovatum), AN4 (Cribbeaturbinispora), AN3(Penicillium capsulatum), showed a sequence similarity of less than 95 % with known organisms in the GenBank. These endophytes could likely be from novel fungal lineages. The biodiversity analysis showed that colonization of endophytic fungi was more significant and more diverse in leaf. Chaetomium sp. Showed the highest colonization (26.9%) frequency, while Colletotrichum sp., Crinipellis sp., Acrophialophora sp., Cribbea sp. showed the least colonization frequency (3.8%).
Conclusion
From the study it is concluded that endophytic fungi show hyper diversity in tropical plants2. Out of 26 isolates maximum endophytes belonging to Ascomycota phylum28. Endophytic fungi belong to Chaetomium sp. were the most abundant fungi occurring in the leaf. With hyper diversity of species and ability to inhabit different environments, Chaetomium sp. might acquire biosynthetic gene clusters, which enables to produce various secondary metabolites for the adaptation of different ecological environments. Chaetomium sp. can be used for futuristic bioactive molecule production exhibiting significant cytotoxic, apoptotic and antioxidant potential29. Fungal endophyte, C. globosum produced chrysin as an alternative resource 30. It is reported that about more than 200 compounds with potential bioactive potential have been isolated from Chaetomium sp31. Recently Chaetomium sp. have taken much attention to be used to manage different economically important diseases. C. globosum has been identified as a potential antagonist of Cochliobolus sativu32.
Acknowledgment
Jawed Shaikhis thankful to the UGC, New Delhi for providing Junior research Fellowship for this research work.
Funding Source
The financial support for this research work was granted from intramural funding of Maulana Azad College of Art, Science and Commerce, Aurangabad.
Conflict of interest
We declare no conflict of interest in the manuscript.
References
- Petrini O, Taxonomy of endophytic fungi of aerial plant tissues, Microbiology of Plant Microbe Interactions 1986;16:580–587.
- Arnold, A. E., Maynard, Z., Gilbert, G. S., Coley, P. D. &Kursar, T. A, Are ropical fungal endophytes hyperdiverse? Ecology Letters 2000;3(4):267–274.
CrossRef - Suryanarayanan, Trichur & Govindan, Venkatesan & Murali, ThokurSreepathy, Endophytic fungal communities in leaves of tropical forest trees: Diversity and distribution patterns, Current Science. 2003;85(5):489-493.
- Schulz, B. & Boyle, C, The endophytic continuum, Mycological Research 2005;109(6):661–686.
CrossRef - Rodriguez R, Redman R, more than 400 million years of evolution and some plants still can’t make it on their own: plant stress tolerance via fungal symbiosis, Journal of experimental Botany 2008;59(5):1109–1114.
CrossRef - Iqbal J, Nelson JA, McCulley RL, Fungal endophyte presence and genotype affect plant diversity and soil-to-atmosphere trace gas fluxes, Plant and Soil 2013;364(½):15–27.
CrossRef - Khan A.L, Waqas M, Hussain J, Al-Harrasi A, Al-Rawahi A, Al-Hosni K, Kim M.-J, Adnan M, Lee I.-J, Endophytes Aspergillus caespitosus LK12 and Phoma sp. LK13 of Moringa peregrina produce gibberellins and improve rice plant growth, Journal of Plant Interactions, 2014;9(1):731–737.
CrossRef - MasroorQadri, Ramesh Deshidi, Bhawal Ali Shah, Kushal Bindu2, Ram A. Vishwakarma1, Syed Riyaz-Ul-Hassan, an endophyte of PicrorhizakurroaRoyle ex. Benth producing menthol, phenyl ethyl alcohol and 3-hydroxypropionic acid, and other volatile organic compounds, World Journal of Microbiology and Biotechnology, 2015;31:1647-1654.
CrossRef - Jouda, J.B.,J.D. Tamokou, C.D. Mbazoa, P. Sarkar, P.K. Bag, and J. Wandji, Anticancer and antibacterial secondary metabolites from the endophytic fungus Penicillium sp. CAM64 against multi-drug resistant Gram-negative bacteria,” African Health Science, 2016;16(3):734-743.
CrossRef - Seetharaman, P.,S. Gnanasekar, R. Chandrasekaran, G. Chandrakasan, A. Syed, M.S. Hodhod, F. Ameen, and S. Sivaperumal, Environmental Science and Pollution Research, 2017;24(26):21272-21282.
CrossRef - Qi, B.,X. Liu, T. Mo, Z. Zhu, J. Li, J. Wang, X. Shi, K. Zeng, X. Wang, P. Tu, I. Abe, and S. Shi, 3,5-Dimethylorsellinic acid derived meroterpenoids from Penicillium chrysogenum MT-12, An endophytic fungus isolated from Huperzia serrata,Journal of Natural Products, 2017;18(10):2699-2707.
CrossRef - Singh, B.; Kaur, A. Antidiabetic potential of a peptide isolated from an endophytic Aspergillus awamori. Journal of Applied Microbiologu. 2016; 120:301–311.
CrossRef - Ferreira, M.C.; Cantrell, C.L.; Wedge, D.E.; Gonçalves, V.N.; Jacob, M.R.; Khan, S.; Rosa, C.A.; Rosa, L.H. Antimycobacterial and antimalarial activities of endophytic fungi associated with the ancient and narrowly endemic neotropical plant Vellozia gigantea from Brazil. Mem. Inst. Oswaldo Cruz 2017; 112:692–697.
CrossRef - Kumar, G.; Chandra, P.; Choudhary, M. Endophytic fungi: A potential source of bioactive compounds. Chem. Sci. Rev. Lett. 2017; 6:2373–2381.
- Sharma R, Kulkarni G, Sonawane MS, Shouche YS A new endophytic species of Chaetomium from Jatropha podagrica. Mycotaxon2013; 124:117–126.
CrossRef - Blanchette RA, Held BW, Abdel-Azeem AM New record of Chaetomium iranianum MF787598 (Chaetomiaceae) for the Egyptian and African mycobiota. MicrobBiosyst Journal 2017;2(2):6–9.
CrossRef - Abdel-Azeem AM, Blanchette RA, Held BW New record of Chaetomium grandeAsgari&Zare (Chaetomiaceae) for the Egyptian and African mycobiota. Phytotaxa 2018;343(3):283–288.
CrossRef - Kalaivani T, Mathew L, Free radical scavenging activity from leaves of Acacia nilotica (L.) Wil. ex Delile, an Indian medicinal tree,” Food and Chemical. Toxicology 2010;48(1):298-305.
CrossRef - Shittu GA, in vitro antimicrobial and phytochemical activities of Acacia nilotica leaf extract, Journal of Medicinal. Plants Research, 2010;4(12):1232-1234.
- Kalaivani T, Rajasekaran C, Suthindhiran K, Mathew L, Free radical scavenging, cytotoxic and hemolytic activities from leaves of Acacia nilotica (l.) wild. ex. delile subsp. indica (benth.) brenan, Evidence Based Complementary Alternative Medicine 2010;2011:274-741.
CrossRef - White, T. J., T. Bruns, S. Lee, and J. W. Taylor, Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, PCR Protocols: A Guide to Methods and Applications, Academic Press, New York, pp. 315-322, 1990.
CrossRef - Darby AC, Chandler SM, Welburn, SC, Douglas AE, “Aphid-symbiotic bacteria cultured in insect cell lines,” Applied Environmental Microbiology, 2005;71(8):4833-4839.
CrossRef - Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, “Basic local alignment search tool,” Journal of Molecular Biology 1990;215(3):403-410.
CrossRef - Tamura K, “Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases,” Molecular Biology and Evolution 1992;9(4):678-687.
- Felsenstein J, Confidence limits on phylogenies: An approach using the bootstrap, Evolution 1985;39(4):783-791.
CrossRef - Kumar S., Stecher G., Li M., Knyaz C., and Tamura K, MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms, Molecular Biology and Evolution 2018;35(6):1547-1549.
CrossRef - Hata K, Futai K, Endophytic fungi associated healthy Pine needle infested by Pine needle gall midge Thecodiplosisjaponensis, Canadian Journal of Botany 1995;73:384–390.
CrossRef - Koukol, O., Kolarík, M., Kolárová, Z., and Baldrian, P, Diversity of foliar endophytes in wind fallen Piceaabies trees, Fungal Diversity 2019;54(1):69–77.
CrossRef - GeethanjaliDhayanithy, KamalrajSubban, JayabaskaranChellilah, Diversity and biological activities of endophytic fungi associated with Catharanthus roseus, BMC Microbiology 2019;19(1):22-36.
CrossRef - SiyaKamat, Madhuree Kumari, KuttuvanValappilSajna&Chellilah. Jayabaskaran, Endophytic fungus, Chaetomium globosum, associated with marine green alga, a new source of Chrysin, Scientific Report 2020;10(1).
CrossRef - Zhang Q, Li HQ, Zong SC, Gao JM, Zhang AL Chemical and bioactive diversities of the genus Chaetomium secondary metabolites. Mini Rev Med Chem 2012;12(2):127–148.
CrossRef - Biswas SK, Aggarwal R, Srivastava KD, Gupta S, Dureja P Characterization of antifungal metabolites of Chaetomium globosum Kunze and their antagonism against fungal plant pathogens. Journal of Biological Control 2012;26(1):70–74.

This work is licensed under a Creative Commons Attribution 4.0 International License.






