How to Cite | Publication History | PlumX Article Matrix
Sounok Sengupta1* ,Ratul Bhowmik2
,Ratul Bhowmik2 , Satarupa Acharjee3
, Satarupa Acharjee3  and Suchandra Sen1
and Suchandra Sen1
1Department of Pharmacology, NSHM Knowledge Campus, Kolkata-Group of Institutions, Kolkata, West Bengal, India.
2Department of Pharmaceutical Chemistry, SPER, Jamia Hamdard, New Delhi, India.
3Department of Pharmaceutical Chemistry, NSHM Knowledge Campus, Kolkata-Group of Institutions, Kolkata, West Bengal, India.
Corresponding Author E-mail: sounok620@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2928
ABSTRACT:
The main objective of this present study was to analyze the anti-inflammatory activity of the compound 1- 3- [3-(substituted phenyl) prop-2-enoyl) phenyl thiourea against inflammation receptors Secretory Phospholipase A2 (sPLA2-X), Cyclooxygenase-2 (COX-2), Interleukin-1 Receptor-associated Kinase 4 (IRAK4), Tumor Necrosis Factor (TNF-alpha) and Inducible Nitric Oxide Synthase 4 using various in-silico techniques. The 3D structures of the receptors were retrieved from Protein Data Bank in PDB format. The ligand molecule was sketched in Chemdraw Ultra v 10.0. The proteins and the ligand molecule were then individually prepared for docking using AutoDock Tools. Docking was performed using AutoDock Vina. Swiss-ADME and Pre-ADMET web servers were used for ADME, drug-likeness, and toxicity analysis. The receptor showing the best binding affinity with our ligand molecule was further analyzed via Molecular Dynamics (MD) Simulations using iMODS web server. The docking results revealed that our ligand molecule showed the best binding affinity with receptor sPLA2-X. The ADME analysis results of our ligand molecule were also good. MD Simulations study showed good results with our ligand- sPLA2-X receptor docked complex. This study revealed that our ligand molecule is a significant inhibitor sPLA2-X and can be further used as a potential therapy against inflammatory disorders.
KEYWORDS: ADME; Chem draw; Inflammation; MD Simulation; Nitric oxide; Phospholipase A2
Download this article as:| Copy the following to cite this article: Sengupta S, Bhowmik R, Acharjee S, Sen S. In-Silico Modelling of 1- 3- [3-(Substituted Phenyl) Prop-2-Enoyl) Phenyl Thiourea Against Anti-Inflammatory Drug Targets. Biosci Biotech Res Asia 2021;18(2). |
| Copy the following to cite this URL: Sengupta S, Bhowmik R, Acharjee S, Sen S. In-Silico Modelling of 1- 3- [3-(Substituted Phenyl) Prop-2-Enoyl) Phenyl Thiourea Against Anti-Inflammatory Drug Targets. Biosci Biotech Res Asia 2021;18(2). Available from: https://bit.ly/38Flo7v |
Introduction
Inflammatory disorders are the most common diseases these days affecting millions of people around the globe. Inflammatory disorders are the cause of the uncontrolled secretion of inflammatory mediators of our body’s cells. Inflammation is a part of the body’s defense system in response to injury or against pathogens. During inflammation, the body synthesizes and secretes several inflammatory mediators which further generate several important cellular effects. Too much secretion of inflammatory mediators leads to several chronic disorders such as cardiovascular disorders, asthma, arthritis, asthma, type 2 diabetes as well as cancer 1,14. To counter inflammatory responses, NSAIDs or non-steroidal anti-inflammatory drugs along with steroidal drugs are mostly prescribed. However, long-term use of these drugs causes several adverse effects such as gastrointestinal toxicity despite demonstrating good anti-inflammatory activities. As of late, various endeavors are made to plan and design drugs that not only would show excellent anti-inflammatory activity but would also show the least side effects 2,15.
A typical thiourea-based chalcone derivative was used as the main drug in this study. Chalcones (trans-1,3-diaryl-2-propen-1-ones) (1) are unsaturated ketones that are made up of two aromatic rings (ring A and B) with a wide range of substituents. Two aromatic rings are connected by an aliphatic three-carbon chain in the chalcone skeleton. A highly electrophilic three-carbon unsaturated carbonyl system connects the two rings of chalcone, forming a linear or nearly planar structure 20. On both aromatic rings, they have conjugated double bonds and a delocalized-electron system. The structural variations of the chalcone rings have resulted in a high degree of diversity, which has proven advantageous for the development of new therapeutic agents, and so chalcones have piqued academic and industrial interest 21,22. The chalcones have been shown to have a wide range of biological actions, including antibacterial, anticancer, cytotoxic, antioxidant, and various other properties 3,4,16.
This study is a follow-up to a previous study that demonstrated the efficacy of a thiourea-based chalcone derivative as an antihyperglycemic drug. The thiourea-based chalcone derivative (1- 3- [3-(substituted phenyl) prop-2-enoyl) phenyl thioureas) was synthesized and biological activity was assessed in a prior study 5. In the previous study, the chalcone-based thiourea derivative, 1- 3- [3-(substituted phenyl) prop-2-enoyl) phenyl thioureas demonstrated anti-hyperglycemic potential as well as protective effects, leading to the restoration of biochemical and antioxidant status as well as the suppression of apoptotic events in the pancreas tissue of STZ-induced diabetic rats. The study also found that diabetic animals’ lipid and renal profiles improved after treatment with this particular chalcone derivative.The main aim of this present study was to check the anti-inflammatory activity of the previously synthesized chalcone derivative with the help of various in-silico tools. In this study, the selected molecule was subjected to molecular docking against several inflammatory receptors such as Secretory Phospholipase A2, Cyclooxygenase-2, Interleukin-1 Receptor-associated Kinase 4, Tumor Necrosis Factor, and Inducible Nitric Oxide Synthase 4 17,18,19. Further, ADMET analysis and molecular dynamics simulation was also performed to check drug likeliness properties and ligand-receptor stability respectively
Materials and Methods
Protein Preparation
The targeted receptors Secretory Phospholipase A2 (PDB-ID: 4UY1), Cyclooxygenase-2 (PDB-ID: 5IKT), Interleukin-1 Receptor-associated Kinase 4 (PDB-ID: 5KX7), Tumor Necrosis Factor (PDB-ID: 2AZ5) and Inducible Nitric Oxide Synthase 4 (PDB-ID: 4NOS) were selected and download in PDB format from protein data bank database (6). The protein molecule was then loaded on AutoDock Tools software (7). Firstly, the extraction of the co-crystallized ligand was done to validate the protein. Immediately after this, protein preparation of the protein was started by removing water molecules, removing chains or heteroatoms not required, repairing missing atoms, the addition of hydrogen atoms, computing charges (Kollman charges) and finally converting it to pdbqt format. Finally, the generation of a grid box was done keeping the co-crystallized ligand at the center. The dimension of the grid box was saved for docking using Autodock Vina as a config.txt file. The co-crystallized ligand was then removed from the prepared protein pdbqt file.
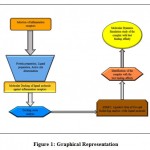 |
Figure 1: Graphical Representation |
Ligand Preparation
On Chemdraw Ultra v 10.0 (Cambridge software), the ligand molecule, 1- 3- [3-(substituted phenyl) prop-2-enoyl) phenyl thiourea, was drawn(Figure 2). To transform a 2D structure to a 3D structure, it was subjected to energy minimization using molecular mechanics (MM2) in Chem3D ultra v 10.0. The minimization process was continued until the root mean square deviation (RMSD) was smaller than.001kcal/mol (8).
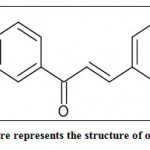 |
Figure 2: This figure represents the structure of our ligand molecule. |
The energy minimized structure was then saved in PDB format using the Chem3D ultra v 10.0 integral option (save as /Protein Data Bank (PDB)). This ligand molecule was then loaded in AutoDock Tools for the further ligand preparation process. Gasteiger charges were added to the ligand molecule. Moreover, non-polar hydrogens were merged, and rotatable bonds were also determined and adjusted. TORSDOF was also used to determine the change in free energy caused by the loss of a torsional degree of freedom upon binding. The prepared ligand molecule was then saved in AutoDockpdbqt format (7).
Molecular Docking using AutoDock Vina
The prepared ligand molecule was then docked individually with the prepared receptors. The docking methodology was performed using AutoDock Vina (9). The results were displayed in terms of binding affinity. The binding affinity represents the binding energy. The binding energy exhibits the extent of binding of the ligand molecule. Furthermore, the best type of configuration would be the one that would bind with its target. The docking results were analyzed using Discovery Studio Biovia 2021(10).
ADMET and Drug Likeliness Analysis
SwissADME and Pre-ADMET web servers were used to predict drug-likeness and ADMET properties of our ligand molecule (11). Lipinski’s rule was used to check whether our ligand molecule was suitable for docking. According to Lipinski’s rule of five, a compound, to qualify as a ligand, should have less than 500 Da molecular weight, high lipophilicity i.e. value of Log P less than five, hydrogen bond donors less than 5, and hydrogen bond acceptors less than 10. Ligand violating any two rules of Lipinski’s is considered unsuitable for docking studies. Other than Lipinski’s rule, physicochemical analysis, as well as Drug-likeliness properties of all the ligand molecules, were also taken into consideration for the drug screening process.
Boiled-EGG Analysis
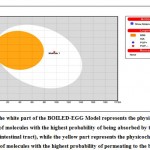
For predicting blood-brain barrier permeability as well as gastrointestinal absorption of our selected phytochemicals, BOILED EGG was used. According to BOILED-Egg plot analysis, compounds found in the yellow region were considered to be having higher blood-brain barrier permeability, whereas compounds found in the white region of the plot were considered to be having higher gastrointestinal absorption properties. The BOILED-Egg plot analysis was performed using the SwissADME webserver (12).
Molecular Dynamics Simulations
It is a computer-based simulation approach used to analyze the physical motions of atoms or molecules. MD simulations can identify a few critical hydrogen bond interactions. MD simulations assist in protein docking and virtual screening advances. The iMODS server was utilized in this work to simulate molecular dynamics. The iMODS service aids in the exploration of normal mode analysis and generate accessible information about routes that may involve macromolecules or homologous structures(13).
Results and Discussion
Molecular Docking Analysis
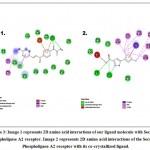
The protein-ligand docking results demonstrated good docking results with the targeted receptors Secretory Phospholipase A2 and Inducible Nitric Oxide Synthase. The chalcone derivative showed the best docking score of -8.6 with the receptor Secretory Phospholipase A2(Figure 3, Figure 4) Other than it showed docking scores of -7.7, -7.9, -5.7, and -8.5 with receptors Cyclooxygenase-2, Interleukin-1 Receptor-associated Kinase 4, Tumor Necrosis Factor and Inducible Nitric Oxide Synthase 4. (Table 1).The co-crystallized ligand of the receptor Secretory Phospholipase A2, 5-(2,5-dimethyl-3-thienyl)-1h-pyrazole-3-carboxamide, previously reported as a SPLA2 inhibitor, was also separately docked with Secretory Phospholipase A2 in order to compare the binding affinity of the chalcone derivative with the co-crystallized ligand. The co-crystallized native ligand, 5-(2,5-dimethyl-3-thienyl)-1h-pyrazole-3-carboxamide, exhibited a docking score of -7.6KJ/mol with receptor Secretory Phospholipase A2, which was quite low in comparison to the binding affinity previously observed between the chalcone derivative and Secretory Phospholipase A2 receptor. Further, the chalcone based thiourea derivative also demostrated ten similar amino acid interaction as that of the native ligand with the receptor Secretory Phospholipase A2. The ten amino acid similar interactions were VAL9, TYR20, PRO17, GLY28, MET21, ILE94, HIS46, ASP47, LEU5, and ALA6 (Figure 3).
Table 1: This table demonstrates the docking score of our ligand molecule with the receptors
| PDB -ID | Protein Name | Ligand | Dock Score |
| 4UY1 | Secretory Phospholipase A2 | Chalcone Derivative | -8.6 |
| 5IKT | Cyclooxygenase-2 | Chalcone Derivative | -7.7 |
| 5KX7 | Interleukin-1 Receptor-associated kinase 4 | Chalcone Derivative | -7.9 |
| 2AZ5 | Tumor Necrosis Factor | Chalcone Derivative | -5.7 |
| 4NOS | Inducible Nitric Oxide Synthase | Chalcone Derivative | -8.5 |
ADMET and Drug Likeness Analysis
The ligand molecule qualified Lipinski’s rule of five. Furthermore, it also demonstrated high gastric retention properties. It exhibited poor skin permeability properties. Furthermore, it also demonstrated high plasma protein binding capacity with a score of 92.096762(Table 2). The toxicity properties retrieved from the Pre-ADMET web server more or less demonstrated the least toxicity indications except for carcinogenicity in mice(Table 3).
Table 2: This table demonstrates the drug-likeness properties of our ligand molecule
| ID | Value |
| BBB | 0.0912741 |
| Buffer_solubility_mg_L | 2.40797 |
| Caco2 | 22.0671 |
| CYP_2C19_inhibition | Non |
| CYP_2C9_inhibition | Non |
| CYP_2D6_inhibition | Non |
| CYP_2D6_substrate | Non |
| CYP_3A4_inhibition | Non |
| CYP_3A4_substrate | Non |
| HIA | 95.335766 |
| MDCK | 2.86401 |
| Pgp_inhibition | Inhibitor |
| Plasma_Protein_Binding | 92.096762 |
| Pure_water_solubility_mg_L | 0.245104 |
| Skin_Permeability | -2.67231 |
| SKlogD_value | 3.62047 |
| SKlogP_value | 3.62047 |
| SKlogS_buffer | -5.11304 |
| SKlogS_pure | -6.10534 |
Table 3: This table demonstrates toxicity parameters of our ligand molecule retrieved from the Pre-ADMET webserver.
| ID | Value |
| algae_at | 0.022025 |
| Ames_test | mutagen |
| Carcino_Mouse | positive |
| Carcino_Rat | negative |
| daphnia_at | 0.0198048 |
| hERG_inhibition | medium_risk |
| medaka_at | 0.00094303 |
| minnow_at | 0.00135928 |
| TA100_10RLI | Negative |
| TA100_NA | Negative |
| TA1535_10RLI | Negative |
| TA1535_NA | Negative |
Boiled-Egg Analysis
The Boiled-Egg plot demonstrated that the chalcone derivative has high gastrointestinal retention properties. Moreover, it also revealed that our drug molecule had no blood-brain barrier permeability property(Figure 5).
Molecular Dynamics Simulations
The Molecular Dynamics simulation results are shown in Figure 6. The complex with the best docking score was considered for MD simulation. Here the docked complex of our ligand with receptor Secretory Phospholipase A2 was considered for MD simulation. Normal mode analysis mobility allows us to analyze the large-scale B-factor and mobility as well as the stability of the molecules. The IMOD server exposed the internal coordinates analysis depending on the protein-ligand structural interactions. IMODs also measure the B-factor and structural deformity and calculate the eigenvalue.
Image A of Figure 6 represents the docked complex of our protein and ligand. Image B of Figure 6 represents the deform ability graph. The deformity graph illustrated peaks in the graph which represent regions in the protein with deform ability. Image C represents the eigenvalue of the complex. The eigenvalue associated with each normal mode represents the motion stiffness. Its value is directly related to the energy required to deform the structure. The lower the eigenvalue, the easier the deformation. Our docked complex demonstrated an eigenvalue of 4.434421e-04. Image D represents the variance plot. The variance plot demonstrates individual variances in red color whereas cumulative variance in green color. Image E represents the elastic map of our docked complex. Each dot in the graph denotes one spring within the respective atoms pair. The dots are colored based on the stiffness where the dark grey dots indicate the stiffer springs and vice versa. Image F represents the covariance map. This map demonstrates the correlation motion between a pair of residues in red color, uncorrelated motion in white color, and anti-correlated motion in blue color. Image G represents the B-Factor graph. The main-chain deform ability or B-Factor is a measure of the capability of a given molecule to deform at each of its residues.
From the molecular dynamics study, it was evident that our complex showed a good amount of deform ability. Furthermore, it also showed a moderately low eigenvalue, suggesting that it could be deformed easily. The variance map exhibited a higher degree of cumulative variances than an individual variance. The elastic network map also produced satisfactory results.
Conclusion
The main aim of our study was to investigate the potency of our chalcone derivative against inflammation mediating receptors Secretory Phospholipase A2, Cyclooxygenase-2, Interleukin-1 Receptor-associated Kinase 4, Tumor Necrosis Factor, and Inducible Nitric Oxide Synthase 4 with the help of computational tools. With the help of in-silico tools, we were able to determine the best binding affinity of our molecule with the targeted receptors. The chalcone derivative showed the best docking score of -8.6 with the receptor Secretory Phospholipase A2 which was further found to be much greater than the binding affinity exhibited by the co-crystallized native ligand with receptor Secretory Phospholipase A2. This indicated potential anti-inflammatory activity of the chalcone derivative against Secretory Phospholipase A2 receptor. Furthermore, the ADME analysis results of our compound were quite positive. Our ligand molecule has good gastric retention as well as good plasma protein binding proteins with very few predicted toxicity parameters. Moreover, the MD simulation study of the docked complex of our ligand with Secretory Phospholipase A2 receptor showed good stability. Hence, we can conclude by saying that our ligand molecule can provide a cure to inflammatory disorders. To find the effectiveness as well as to propose the exact mechanism, in-vitro studies can be encouraged on our compound to understand the exact mechanism and potential cure for inflammatory disorders.
Acknowledgment
The research work was self-financed and the study was carried out totally by all authors so there is no need for acknowledgment.
Funding Source
There is no funding source.
Conflict of Interest
There is no conflict of interest.
References
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–7.
CrossRef - Tabas Ira, Christopher G. Anti-Inflammatory Therapy in Chronic Disease: Challenges and Opportunities. Science (80- ). 2013;339(6116):166–72.
CrossRef - Tekale S, Mashele S, Pooe O, Thore S, Kendrekar P, Pawar R. Biological Role of Chalcones in Medicinal Chemistry. Vector-Borne Dis – Recent Dev Epidemiol Control. 2020;
CrossRef - Rozmer Z, Perjési P. Naturally occurring chalcones and their biological activities. Phytochem Rev. 2016;15(1):87–120.
CrossRef - Acharjee S, Maity TK, Samanta S, Mana S, Chakraborty T, Singha T, et al. Antihyperglycemic activity of chalcone based novel 1-{3-[3-(substituted phenyl) prop-2-enoyl] phenyl} thioureas. Synth Commun [Internet]. 2018;48(23):3015–24. Available from: https://doi.org/10.1080/00397911.2018.1539178
CrossRef - Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Vol. 28, Nucleic Acids Research. 2000. p. 235–42.
CrossRef - Morris GM, Ruth H, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. Software news and updates AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–91.
CrossRef - Cousins KR. Computer review of ChemDraw ultra 12.0. J Am Chem Soc. 2011;133(21):8388.
CrossRef - Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009;31(2):455–61.
CrossRef - Visualizer DS. v4. 0.100. 13345. In: Accelrys Software Inc. 2005.
- Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(March):1–13.
CrossRef - Daina A, Zoete V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem. 2016;11(MAY):1117–21.
CrossRef - Lopéz-Blanco JR, Garzón JI, Chacón P. iMod: Multipurpose normal mode analysis in internal coordinates. Bioinformatics. 2011;27(20):2843–50.
CrossRef - Chen H, Knerr L, Åkerud T, Hallberg K, Öster L, Rohman M, et al. Discovery of a novel pyrazole series of group X secreted phospholipase A2 inhibitor (sPLA2X) via fragment based virtual screening. Bioorganic Med Chem Lett [Internet]. 2014;24(22):5251–5. Available from: http://dx.doi.org/10.1016/j.bmcl.2014.09.058
CrossRef - Orlando BJ, Malkowski MG. Substrate-selective inhibition of cyclooxygeanse-2 by fenamic acid derivatives is dependent on peroxide tone. J Biol Chem [Internet]. 2016;291(29):15069–81. Available from: http://dx.doi.org/10.1074/jbc.M116.725713
CrossRef - Hanisak J, Seganish WM, McElroy WT, Tang H, Zhang R, Tsui HC, et al. Efforts towards the optimization of a bi-aryl class of potent IRAK4 inhibitors. Bioorganic Med Chem Lett [Internet]. 2016;26(17):4250–5. Available from: http://dx.doi.org/10.1016/j.bmcl.2016.07.048
CrossRef - He MM, Smith AS, Oslob JD, Flanagan WM, Braisted AC, Whitty A, et al. Small-Molecule Inhibition of TNF- a. 2005;310(November):1022–5.
CrossRef - Fischmann TO, Hruza A, Niu X Da, Fossetta JD, Lunn CA, Dolphin E, et al. Structural characterization of nitric oxide synthase isoforms reveals striking active-site conservation. Nat Struct Biol. 1999;6(3):233–42.
CrossRef - Čopič A, Vučemilo N, Gubenšek F, Križaj I. Identification and purification of a novel receptor for secretory phospholipase A2 in porcine cerebral cortex. J Biol Chem. 1999;274(37).
CrossRef - Jantan I, Bukhari SNA, Adekoya OA, Sylte I. Studies of synthetic chalcone derivatives as potential inhibitors of secretory phospholipase A2, cyclooxygenases, lipoxygenase and pro-inflammatory cytokines. Drug Des DevelTher. 2014;8.
CrossRef - Li S, Jiang M, Wang L, Yu S. Combined chemotherapy with cyclooxygenase-2 (COX-2) inhibitors in treating human cancers: Recent advancement. Vol. 129, Biomedicine and Pharmacotherapy. 2020.
CrossRef - Zarghi A, Arfaei S. Selective COX-2 inhibitors: A review of their structure-activity relationships. Vol. 10, Iranian Journal of Pharmaceutical Research. 2011.

This work is licensed under a Creative Commons Attribution 4.0 International License.