How to Cite | Publication History | PlumX Article Matrix
Ashwini Talakayala1,2 , Veerapaneni Bindu Prathyusha1
, Veerapaneni Bindu Prathyusha1 , Dhanasekar Divya 1
, Dhanasekar Divya 1 , Srinivas Ankanagari2
, Srinivas Ankanagari2 and Mallikarjuna Garladinne1*
and Mallikarjuna Garladinne1*
1Plant Molecular Biology laboratory, Agri Biotech Foundation, Rajendranagar, Hyderabad-500 030.
2Department of Genetics, Osmania University, Hyderabad-500 007.
Corresponding Author E-mail: galradinnemarjun@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2932
ABSTRACT: Mungbean yellow mosaic virus (MYMV) causes massive crop losses in green gram. MYMV is a member of begomovirus with bipartite genome comprising DNA-A and DNA-B components, which is transmitted by whiteflies. Cloning and preparation of infectious clone is very much essential for screening germplasm or transgenic material of pulse crops since viruliferous whiteflies may not be available throughout the year. In the current work, we have amplified rolling circle mediated viral genome of MYMV using Φ29 DNA polymerase. The amplified products was digested and cloned into the plant expression vector pCAMBIA2301.The cloned constructs was then transformed into Agrobacterium LBA4404 through freeze thaw method. Further, three viral transmission techniques including mechanical rubbing, Agroinfiltration and Agroinoculation, were employed for assessing the mosaic symptoms in green gram. The molecular confirmation through polymerase chain reaction (PCR) indicated that the yellow mosaic symptoms were formed due to infectivity of MYMV in the green gram.
KEYWORDS: Agroinfiltration; Agroinoculation; Φ29 DNA polymerase; Yellow mosaic disease
Download this article as:| Copy the following to cite this article: Talakayala A, Prathyusha B. V, Divya D, Ankanagari S, Garladinne M. Molecular Cloning of MYMV Genome and Infectivity of Yellow Mosaic Virus in Green Gram using Different Viral Transmission Tools. Biosci Biotech Res Asia 2021;18(3). |
| Copy the following to cite this URL: Talakayala A, Prathyusha B. V, Divya D, Ankanagari S, Garladinne M. Molecular Cloning of MYMV Genome and Infectivity of Yellow Mosaic Virus in Green Gram using Different Viral Transmission Tools. Biosci Biotech Res Asia 2021;18(3). Available from: https://bit.ly/2ZyGaoe |
Introduction
Yellow Mosaic Disease in green gram is caused by Mungbean Yellow Mosaic Virus (MYMV), devastating large acres of crops, particularly in tropical and subtropical countries1. The MYMV belongs to the Geminiviridae family and two isolates of MYMV were evolved in Indian sub-continent. The MYMV possesses a circular and single-stranded DNA genome encapsulated with icosahedral particles. Generally, Begomovirus have either monopartite DNA-A or DNA-B (Concatomeric 2.6 or 2.7 kb) or bipartite (Concatomeric 5.2 kb) circular ssDNA genome. The DNA-A genome has five open reading frames (ORFs), encoding for functional proteins i.e., AV1 coat protein (CP), AC1 viral replication initiation protein (Rep), AC2-Transcriptional activator (Trap), AC3-replication enhancer (Ren), and AC4. The genome also has two ORFs in DNA-B encoding i.e., movement or mobility protein (MP) and a nuclear shuttle protein (NSP)2. The predominant geminiviruses are causative entities in leguminous species including greengram, black gram, french gram, pigeon pea, and soybean3. MYM virus is transmitted by whitefly, Bemisia tabaci Genn. (Hemiptera: Aleyrodidae). It is a polyphagous insect, completes its life cycle in less than 2 weeks to more than 10 weeks depending on temperature and host plant where it has greater ability to transmit disease4.
Geminiviruses are circular single-stranded (ss) DNA plant pathogens that can replicate double- stranded (ds) DNA via rolling circle mode of amplification (RCA) in host plants5. RCA is a process in which a circular DNA or RNA molecule is replicated in one direction through strand displacement activity of Φ29 DNA polymerase. First DNA molecule is displaced by newly synthesized DNA and releases the single-stranded DNA (ssDNA). The primer enzymatic extension combined with strand displacement generates a long ssDNA complementary to the DNA template. RCA is recognized as an important diagnostic tool to amplify the complete genome of viruses artificially under in vitro conditions. This technique is used to detect many ss, ds DNA viruses infecting different crops6. The double stranded circular papilloma viral genome sample of infected leaf tissue was efficiently amplified using Φ29 DNA polymerase via RCA technique7, 8. Similarly, Inoue-Nagata et al.9 used cloning of single circular DNA method in tomato for Tomato Chlorotic Mottle Virus.
The whiteflies act as natural vectors carrying viruses that spread viral diseases10. The host phloem cells are used by virus to enter and their viral aggregates pass on through it. The symptoms appear within two days as a scattered yellow spot on the young leaves turning to mosaic appearance, infected plant pods size decreases, leaf yellowing decreasing the efficiency of photosynthesis and causing a severe crop losses11. The insect would not persist in all seasons; hence, researchers are encountering issues with screening germplasm and assessing resistant varieties. In vitro cloning of the complete viral genomes through RCA is possible to develop screening techniques like Agro-inoculation, Agroinfiltration. Cloning a viral genome in a suitable vector is the possible solution for screening and challenging the testing material in the laboratory without any climatic barrier and escaping the transfer of viral components.
Viral transmission studies help in understanding inplanta gene – gene interaction, gene expression and functional analysis. Further, the Agroinoculation of viral genome into plants would be useful for understanding viral replication, assembly and their movement. Similar technique was followed using soyabean isolate to infect in green gram and blackgram12. In another study the tobacco leaves were infiltrated with Agro-clone for multiplication and propagation of Potato virus X (PVX)13. Madhuitha et al 14 has evaluated Vigna germplasm was to detect the level of resistance/susceptibility against yellow mosaic virus.
Agroinoculation and Agroinfiltration methods were first applied for the of both viral DNA/RNA genomes such as Cauliflower Mosaic Virus (CaMV) in turnip plant Maize Streak Virus (MSV) in maize15, 16. Vaghchhipawala et al. 17 reported that by using virus-induced gene silencing (VIGS) method to study plant-virus interactions, functional analysis of viral genomes and also for genetic screening with forward and reverse approaches. These techniques are applicable to investigate transient gene expression effects, analyze its protein localization and protein-protein interactions in plant-pathogen studies. Agroinfiltration method was used for delivering viral vectors carrying Tobacco Mosaic Virus (TMV) and Potato Virus X (PVX) in leaf part or whole plant for gene expression studies17. Through Agroinfiltration method, sGFP tagged viruses of Tomato Torrado virus was inoculated in tomato and tobacco for studying molecular determinants during viral movement for analyzing plant-virus interactions.18 A cDNA clone of tomato mottle moasic virus (ToMMV) was used for agroinfiltration of tomato leaves, where pathogenicity and virus host interactions were studied through electron micrographs19.
In present study, we report a simple procedure for construction of agro-infectious genomic clones of MYMV. The complete genome of MYMV was amplified using Φ 29 DNA polymerase and digested genome was cloned in pCAMBIA2301 plant expression vector. This vector was used for Agroinfiltration and Agro-inoculation studies in greengram. The yellow mosaic symptoms in infected samples were assessed and screened through PCR using genes specific markers encoding DNA-A and DNA-B in plants. .
Materials and methods
Extraction of genomic DNA from infected leaves of greengram
The greengram plants exhibiting mosaic symptoms with irregular green and yellow spots were collected from greengram fields in Acharya N. G. Ranga Agricultural University, LAM farm, AP, India (Fig 1A). Genomic DNA was extracted from infected leaf samples and control plants by following CTAB method. Briefly, infected leaves were ground with CTAB buffer (10 mM EDTA, 100 mM Tris, 1.4-2.0 M NaCl, 2% CTAB, 2-5% ß-mercaptoethanol). The leaf slurry incubation was done at 60º C for 30 min, equal volumes of Phenol: Chloroform : Iso-amylalcohol (25: 24: 1) was added to the samples supernatant and centrifuged for 10 min at 13,000 rpm. The supernatant was then added with Chloroform: Iso-amylalcohol (24: 1) followed by centrifugation. Further the supernatant was precipitated with the addition of 100 % isopropanol and 7.5 M Sodium acetate. Finally, the pellet was washed with 70 % ethanol and DNA pellet was dissolved in sterile water 20. The extracted DNA was qualitatively measured by resolving in 0.8 % agarose gel, then purity was analyzed at OD of 260 and 280 nm by NanoVue spectrophotometer (GE healthcare, USA).
Rolling circle amplification of MYMV genome
The genomic DNA from infected and control samples was subjected to RCA using Φ 29 DNA polymerase (Thermo Scientifics, USA) according to the instructions given by manufacturer. Briefly, reaction mixture was prepared by the addition of 100 ng of RCA DNA, 500 µM Exo- Resistance Random primer and adjusting reaction volume for 10 µl with water. Following with incubation of the reaction mixture was done at 5 min at 95ºC and chilled on ice for 2 min. To this reaction mixture, 10 mM dNTPs and 1 µl Φ29 DNA polymerase (10 U/µl) and Pyrophosphate inorganic (0.1 U/µl) was added and the final volume was made up to 15 µl – 20 µl based on DNA concentration. The above mixture was then incubated at 30 ºC for 18 hrs and the reaction was terminated by keeping at 65 ºC for 10 min.
Confirmation of presence of DNA-A and DNA-B components in MYMV genome
To confirm whether the RCA possessing, DNA-A and B, was processed using gene specific primer sets such as AC1, VCP2 (AV1), BC1 and BV1 through PCR. The reaction performed in 10 μl containing 10 x PCR reaction buffer, 2.5 mM dNTPs, 2.5 mM MgCl2, each primer 10 pmols, 2.5 U/μl Taq DNA polymerase (Takara) and 100 ng RCA-DNA. The primer details are given in (Table 1). The PCR program was followed as 94ºC for 5 min for initial denaturation; 94 ºC for 40s; 54 ºC for 40s for annealing of AC1, VCP2 (AV1), BC1 and BV1, extension at 72ºC for 40s and 72ºC for 7 min for final extension. After the amplification, the PCR products were resolved on 1% agarose gel in 1x TAE buffer at 100v through electrophoresis. The gel was analyzed with gel documentation system (Syngene, USA).
Table 1: Primers used for amplifying selected regions and full-length DNA-A and DNA-B components of yellow mosaic viruses infecting green gram.
| MYMV
Genome |
Gene | MYMV isolates of Vigna Species
Accession No |
Primers: Nucleotide Sequence | Primer
Size |
Annealing
Temp |
Product
Size (kb) |
|
DNA –A |
AC1 | AB017341.1 | FP: 5’ ATGCCTAGACTCGGTCGTTTTG 3’ | 22 bp |
54 ℃
|
0.7 kb |
| RP: 5’ CGTGCGACTATCGCCTTCAATC 3’ | 22 bp | |||||
| AV1
(VCP2) |
DQ400848.1 | FP: 5’ GCCAAAGCGGAATTACGA 3’ | 18 bp |
0.7 kb |
||
| RP: 5’ GCCTCTTGGTGGTTGTAAAC 3’ | 20 bp | |||||
|
DNA-B |
BC1 | AJ439059.1 | FP: 5’ATGGAGAATTATTCAGGCGCAG 3’ | 22 bp |
0.8 kb |
|
| RP: 5’ TTACAACGCTTTGTTCACATTG 3’ | 22 bp | |||||
| BV1 | DQ400849.1 | FP: 5’ ATGTTTAACCGCAATTATCGCA 3’ | 22 bp |
0.7 kb |
||
| RP: 5’ TTATCCCACGTATTTCAATTCA 3’ | 22 bp |
Construction of pCAMBIA2301 vector with MYMV genome RCA product
RCA products were digested with Bam HI and Hind III by following manufacturer’s instructions (Thermo Scientific, USA). RCA products were digested individually with Bam HI and Hind III using about 2 µg of RCA product, 10 µl buffer, 10 U/µl of Bam HI and Hind III restriction enzyme, adjusting reaction volume with water to 50-100 µl, incubated for 25 min at 37ºC and were inactivated by an enzyme at 85ºC for 10 min. Further, 0.8% Agarose gel was used to resolve the digested products. The digested DNA fragments were excised from the gel was eluted with Qiagen Gel Extraction kit (Qiagen, USA). The eluted DNA fragments (300 ng) were ligated with pCAMBIA2301 using T4 ligase (Thermo scientific, USA) individually (Fig 1B).
Coli and Agrobacterium transformation
The ligated products that contain pCAMBIA2301 vector and MYMV genome (2.6 kb and 2.7 kb) fragments separately was transferred into E. coli (DH5α) by heat shock method21. About 10 µl ligated product was added into competent DH5α cells. The mixture was incubated on ice for 5 min, and given heat shock at 42°C for 90 s and incubated on ice for 1 min. To the mixture, 1 ml of LB broth was added and incubated for 1 hr at 37°C, 180 rpm in the incubator. After the incubation, 50 µl cell suspension was spread onto LB agar medium supplemented with kanamycin (50 mg/L) and incubated at 37°C for overnight (Fig 2A). Then the colony PCR was performed for detecting the cloned fragments such as DNA-A i.e., AC1, AV1 (VCP2), and DNA-B i.e., BC1, BV1 using gene specific primers. Plasmid DNA was isolated from positive colonies using Qiagen Plasmid isolation kit (Qiagen, USA). The sequencing of amplified fragments was done at DNA sequencing facility (Eurofins Genomics India Pvt, Ltd, Bangalore). Analysis of sequenced data of MYMV DNA was carried out by multiple sequence alignment.
The two plasmids DNA-A and DNA-B were then transferred separately into Agrobacterium through freeze thaw method. About 1 µg of plasmid DNA was added into 200 µl of Agrobacterium LBA4404 cells, and snap freezed in liquid nitrogen, 1 ml of LB broth was and incubated at 28°C for 4 hr. The suspension was then plated on LB agar plates supplemented with streptomycin (50 mg/L) kanamycin (50 mg/L), and rifamycin (20 mg/L) followed by incubation at 28°C for 48 -72 hr. Plasmid DNA was isolated from positive colonies using Qiagen Plasmid isolation kit (Qiagen, USA) and glycerol stocks were prepared and used for Agrobacterium transmissions.
Viral transmission studies in green gram
Rubbing
Mechanical transmission of MYM in green gram was performed using rubbing method. The sap was prepared from MYMV infected leaves of green gram by macerating in the cold sodium phosphate buffer (300 mg of powdered tissue with 1.5 ml of sodium phosphate buffer) and centrifuged at 4°C at 13,000 for 5 min. About 100 µl of supernatant was rubbed on the adaxial surface of 3-4 fully expanded leaves per plant. Symptoms were scored after 2 days of rubbing. The genomic DNA was extracted from infected leaves and analyzed through the PCR using AC1 gene specific primers.
Agro-infiltration:
Agrobacterium containing pCAMBIA2301 vector harbouring DNA-A and DNA-B was grown till the OD600 reaches 0.4 – 0.6 at 28°C for 48 hrs. Then, the cultures was centrifuged at 5,000 rpm and each culture was diluted in three volumes of Agro-infiltration medium (10 mM MgCl2, 10 mM MES, pH 5.6) until culture OD600 reached to 0.5. Further, culture was incubated at room temperature with gentle shaking for 1-2 hrs. Both cultures were mixed together along with 100 µM acetosyringone and incubated for 2 hrs for vir gene induction13. Culture was infiltrated on abaxial surface of leave without any needle to infiltrate leaves of 4 to 5-week-old plants of greengram. After post infection, occurrence of yellow mosaic symptoms (4 to 8 days), the genomic DNA was extracted from infected leaves and analyzed through the PCR using AC1 gene specific primers (Figure 3).
Agro-inoculation
The green gram seeds were surface sterilized with 70% ethanol and imbibed in water for overnight. The seeds were pricked with fine syringe and inoculated with medium along with 100 µM acetosyringone and seeds were maintained at 28°C for 3 hrs and excess culture was removed and seeds were sown in soil. Post 15-21-days of infection, the MYMV symptoms were phenotypically characterized and the symptoms were recorded in the trifoliate leaves. The uninoculated seeds of each line were maintained as control.
Results and Discussion
Detection and Confirmation of DNA-A and DNA-B components of MYMV genome
Yellow mosaic disease (YMD) of green gram and black gram is economically very important and it is caused by two species of virus, MYMV and MYMIV and are transmitted by white flies. Incidence of this disease leads to cent percent yield loss of the crop. Hence it is necessary to develop the MYMV resistant varieties through Marker Assisted Breeding. To identify the new sources of resistant variety, we need to screen the germplasm against MYMV. Screening under the field conditions was always influenced by the different factors like i.e., environmental changes, whitefly genotypes etc.. Advancement in the molecular biology provides us three different method of screening methods: Rubbing method, Agroinfiltration and Agroinoculation (Aguilar et al13 ; Usharani et al. 200623 ; Biswas and Varma 24 ;Mandal et al.25). In the present study, we have followed these three techniques for green gram screening against YMD. Green gram plants with typical characteristics of mosaic symptoms including green and yellow patches (Fig 1A) with lesser flowers and pods in the fields were chosen for this experiment. The infected and control leaf samples were collected for DNA extraction. About 20 µg of DNA was extracted from those infected leaves. The DNA contains both plant and viral genomic DNA. The purity of the DNA was analyzed by NanoVue at OD 260/280 with 1.8. The complete genome of MYMV was amplified from 200 ng of genomic DNA as a template by using Φ29 DNA polymerase (Fig 1B). In this method, the DNA fragments amplifies as mini circles through rolling circle amplification by random priming for generating high concentration of concatermerized DNA Dean et al.22 . Then the RCA product was partially digested with Bam HI and Hind III, obtained a fragments with a size of 2.6 kb and 2.7 kb each. The digested product was confirmed by amplifying coat protein by using gene specific primers and obtained 0.7 kb fragment (Fig 1C). Similar to our work, Kumar et al.26 reported 2.6 kb and 2.7 kb fragments of DNA A and DNA B isolated from cowpea against MYMV. Similarly, in the other report Jyothsna et al 27 reported 2.7 kb and 2.6 kb of viral genomes isolated from Rhynchosia minima weed.28 In contrast, Haq et al.28 reported 2 kb and 1.5 kb fragments of DNA-A and B from blackgram.. Based on the reports to avoid the confusion we have amplified the DNA-A and DNA-B with reported specific primers and sequenced.
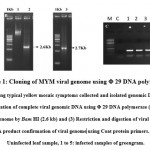 |
Figure 1: Cloning of MYM viral genome using Φ 29 DNA polymerase |
RCA products cloning in pCAMBIA2301 expression vector and Agrobacterium transformation
Geminiviruses, damages many crop on several countries. To control the spread of the virus demands efficient diagnostic tools like specific antibodies, PCR and RFLP (Briddon and Markham29). Later Dean et al.22 provided the feasible procedure by the uses of Bacillus subtilis bacteriophage Phi29 polymerase. By using this DNA polymerase we can able to do polymerase and stand displacement activity through RCA mechanism. Through this RCA approach we can detect and characterize geminiviruses and related subgenomic components (Haible et al.1). In the present study, we have used the RCA mechanism to characterize YMD in green gram. The digested DNA fragment was cloned in the Bam HI site of pCAMBIA2301 using T4 DNA ligase and another DNA fragment was cloned in the Hind III site of pCAMBIA2301 using T4 DNA ligase. Then, two plasmids were transferred into Agrobacterium for further viral transmission studies. The Agro colonies appeared in 48 hr of incubation (Fig 2A). The colonies were confirmed by PCR using a different set of primers including AC1 (replication protein), BC1 (mobility protein) and BV1 (shuttle protein), which showed expected amplification products of 0.7 kb, 0.8 kb and 0.7 kb respectively (Fig 2B and 2C). All these fragments were sequenced by Eurofins Genomics, Bangalore. About 97% of DNA-A sequence was matched with accession no DQ400847.1 isolated from black gram. This sequence also matches 96-97% with the DNA-A of another crop like soyabean, French Bean, green gram infected with YMD. In case of DNA-B the sequences matched 97% with the accession no AJ439059.1. reported from mungbean. DNA-B sequences matched only with mungbean not with any other crops . Based on the blast and Clustal w analysis, we confirmed our isolated clone belongs to YMD DNA-A and DNA-B and used for agrotrans formation. . The confirmed Agrobacterium LBA4404 clone with MYMV genome was used for further virus transmission studies.
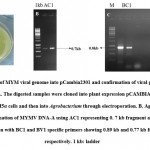 |
Figure 2: Cloning of MYM viral genome into pCambia2301 and confirmation of viral genes using gene specific primers. |
Mechanical transmission with MYMV maintained Agrobacterium clones
Transmission may be due to rubbing of plants with liberated virus from damaged hair cells or epidermal cells 30 . The virus is transmitted by rubbing the viral sap on to the adaxial side of trifoliate leaves of greengram (Fig 3A). The greengram is highly susceptible to MYMV. The severe yellow mosaic symptoms appeared on greengram leaves in 7-10 days post infection (Fig 3B). Further, the infected and control plant were used for detecting viral particle accumulation in the plants through PCR. The PCR results indicated that the AC1 fragment with 0.7 kb was amplified in all infected samples. However, uninfected control did not showed any symptoms and failed to show amplification with AC1 primers(Fig 3C). . About 90±1 % of 10 leaflets rubbed with sap extracted from MYMV infected greengram plants in triplicates, developed yellow mosaic symptoms after 5 days of post infection. During this period, it allowed the virus particles penetrate the leaf surface. Aguilar et al.13 reported that the mechanical rubbing of PVX sap inoculum on the surface of Nicotiana benthamiana allowed penetration and propagation of viral particles and the symptoms appeared in 7 days of post infection. Similarly, rubbing method was reported for transmission of sugarbeet virus in Beta vulgaris and Spinacia oleracea crops (Kassanis31). Another Begomovirus, Curly top virus which affects the different crops i.e., tomatoes, beans, cassava, squash and cotton also transmitted by rubbing method (Saad et al.32). Singh and Awasthi 33 reported different artificial mechanical transmission of virus like TMV, CMV, TSV, Okra mosaic virus, Papaya ringspot virus, Cowpea mosaic virus and alfalfa mosaic virus in tobacco, tomato, sunflower, okra, papaya, cowpea and potato (Sacrista´n et al.34; Jalender et al.35; Sundaresha et al.36; Givord and Hirth 37 ; Gonsalves 38 ; Surekha et al.39; El-Abhar et al. 40).
 |
Figure 3: Viral transmission studies using mechanical rubbing. |
Agroinfiltration using MYMV Agrobacterium clone
Agro culture was infiltrated on the abaxial side of the trifoliate stage of greengram. The severe yellow mosaic symptoms were observed in 21 days of post infection (dpi) in greengram leaves. None of the symptoms observed in control leaves. The viral DNA was detected in Agroinfiltrated leaves using AC1 gene specific primers. The expected amplicon size of 0.7 kb was obtained in two out of three symptomatic plants (Fig 4C). However, the uninfected control plants didn’t show any amplification. Zhang et al.41 constructed cDNA clone of Wheat yellow mosaic virus (WYMV) genomic RNA and used Agroinfiltrated for viral multiplication and systemic infection in tobacco and wheat. Interestingly synergic effect was detected between WMYV and Chinese wheat mosaic virus (CWMV).
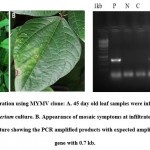 |
Figure 4: Agro infiltration using MYMV clone |
Liu et al 42 reported that through agroinfiltration tobacco rattle virus (TRV) was transmitted in tomato and studies the functional analysis in detail (Liu et al., 42,43,44; Dong et al., 45 ; and Anand et al., 46 ). In another study, Gao et al., 47 reported that agroinfiltration studies had typical mosaic symptoms in J.curcas with Jatropha curcas mosaic disease and used for screening the cultivars to identify the virus resistant plants. Wieczorek et al.,48 developed a construct of infectious cones of tomato torrado virus and transmitted to plants through agroinfiltration.. Similarly, Usharani et al., 49 studied AV promoter expression via., infiltration in tobacco and different legumes by a reporter gene i.e., GUS.
Transmission of MYMV using Agro-inoculation technique
Agroinoculated plants were assessed for yellow mosaic symptoms in Green gram and Black gram. For this experiment, LGG460 (green gram) and LBG685 (black gram) lines were subjected to Agro-inoculation in five replications. Severe yellow mosaic symptoms were observed in 14 days after inoculation in trifoliate leaves of black gram and green gram (Fig 5D and 5F), however, no symptoms were developed in the control uninoculated plants. The genomic DNA was extracted from the leaves showing yellow mosaic symptoms. The PCR confirmation was done for the viral DNA presence i.e., MYMV AC1 gene and all inoculated samples showed expected amplification of 0.7 kb fragment (Fig 5H). MYMV symptoms were appeared 90% of green gram and 84.6% in black gram plants inoculated with Agro infectious clone. However, the resistant lines in green gram and black gram have shown minimal symptoms (Table 2). Jacob et al 50 reported MYMV which infects blackgram, mungbean and soyabean. Usharani et al.12 demonstrated viral DNA presence in symptomatic plants agro-inoculate. Asymptomatic of resistant genotypes compared to control plants of were compared with controls through PCR. Advantage of agroinoculation is to observe the uniform symptoms in all infected leaves, which would make easier to compare with control plants24. MYMV symptoms are analysed based on the scattered appearance of yellow-color spots on young leaves which turns into mosaic pattern11. Sudha et al.51 screened germplasms of mungbean to identify the resistant/susceptible against MYMV and analysed its viral load by PCR with coat protein gene primers.
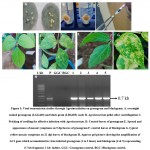 |
Figure 5: Viral transmission studies through Agroinoculation on greengram and blackgram. |
Table 2: PCR screening based on appearance of symptoms in black gram and green gram after challenging with MYMV through Agro inoculation.
| Variety | No of seedling inoculated | No of seedling germinated | No of plants showed YMD symptoms | No of plants showed AC1 amplification | Percentage of plants observed with symptoms |
| LGG460 (Gg-S) | 15 | 10 | 9 | 10 | 90% |
| GG150 (Gg-R) | 15 | 14 | 1 | 1 | 7.1% |
| LBG685(Bg-S) | 15 | 13 | 11 | 11 | 84.6% |
| PU31(Bg-R) | 15 | 15 | 1 | 1 | 6.6% |
Agroinoculation studies were used to screen Tomato yellow leaf curl virus in Sweet pepper, and Pepper yellow leaf curl virus in capsicum (Kil et al., 52; Koeda et al., 53). In another study, Overcoming a resistance against Tomato yellow leaf curl virus in Lycopersicon species was observed by Kheyr et al.54.
Conclusion
The MYMV genome was amplified using Φ29 DNA polymerase from infected samples of green gram. The cloned MYMV construct is very useful in bioassay experiments for screening different genotypes of green gram, black gram and other crops. These viral transmission tools would be employed for screening the CRISPR mediated yellow mosaic virus resistance in pulse crops.
Acknowledgement
We sincerely acknowledge the funding support of Council of Scientific and Industrial Research (CSIR) with No. 38(1450)/18/EMR II. We thank Executive Director and Agri Biotech Foundation for encouragement and support
Conflicts of interest
The authors declare no conflicts of interest
Funding Sources
The grants funded by Council of Scientific and Industrial Research, INDIA under Extra Mural Research scheme to Dr. G. Mallikarjuna, Agri Biotech Foundation, Hyderabad, India. Ref no. 38(1450)/18/EMR II.
References
- Haible D., Kober S. and Jeske H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J .Virol. Methods. 2006; 135, 9-16
CrossRef - Rojas M.R., Hagen C., Lucas W.J. and Gilbertson R.L. Exploiting chinks in the plant’s armor: evolution and emergence of geminiviruses. Rev. Phytopathol. 2005; 43, 361-394.
CrossRef - Fauquet C.M. and Stanley J. Geminivirus classification and nomenclature: progress and problems. Appl. Biol. 2003; 165-189.
CrossRef - Nariani T.K. Yellow mosaic of mung (Phaseolus aureus ). Ind. Phytopathol. 1960; 13:24-29
- Jeske H. Geminiviruses. In: TT 2009; 185-226.
CrossRef - Bhat A.I. and Rao G.P. Rolling Circle Amplification (RCA), In Characterization of Plant Viruses- Methods and Protocols (Humana, New York: Springer). 2020; 377–382.
CrossRef - Rector A., Tachezy R., Van Doorslaer K., MacNamara T., Burk R.D., Sundberg J.P. and Van Ranst M. Isolation and cloning of a papillomavirus from a North American porcupine by using multiply primed rolling-circle amplification: the Erethizon dorsatum papillomavirus type 1. Virology, 2005; 331, 449-456.
CrossRef - Stevens H., Rector A. and Van Ranst M. Multiply primed rolling-circle amplification method for the amplification of circular DNA viruses. Spring. Harb. Protoc. 2010; pdb.prot5415.
CrossRef - Inoue-Nagata A.K., Albuquerque L.C., Rocha W.B. and Nagata T. A simple method for cloning the complete begomovirus genome using the bacteriophage φ29 DNA polymerase. Virol. Methods. 2004; 209-211.
CrossRef - Selvi R., Muthiah A.R., Manivannan N., Raveendran T.S., Manickam A. and Samiyappan, R. Tagging of RAPD marker for MYMV resistance in mungbean (Vigna radiata (L.) Wilczek). J. Plant Sci. 2006; 277-280.
CrossRef - Malathi V.G. and John P. Mungbean yellow mosaic viruses. Desk encyclopedia of plant and fungal virology. Press, London. 2009; 217-226.
CrossRef - Usharani, K.S., Surendranath, B., Haq, Q.M. and Malathi, V.G., 2005. Infectivity analysis of a soybean isolate of Mungbean yellow mosaic India virus by agroinoculation. Gen. Plant. Pathol.2005; 230-237.
CrossRef - Aguilar E., del Toro F.J., Chung B.N., Canto T. and Tenllado F.,. Infection of Nicotiana benthamiana Plants with Potato Virus X (PVX). Bio-Protoc. 2016; 6: e2063
CrossRef - Madhumitha B., Karthikeyan A., Mathivathana M.K., Aiyanathan K.E.A. and Sudha M. Evaluation of Vigna germplasms for resistance to mungbean yellow mosaic virus using agro inoculation. J. Plant. Breeding. 2019; 692-698.
CrossRef - Grimsley N., Hohn B., Hohn T. and Walden R. “Agroinfection,” an alternative route for viral infection of plants by using the Ti plasmid. Natl. Acad. Sci. 1986; 3282-3286.
CrossRef - Grimsley N., Hohn T., Davies J.W. and Hohn B. Agrobacterium-mediated delivery of infectious maize streak virus into maize plants. Nature. 1987; 177-179.
CrossRef - Vaghchhipawala Z., Rojas C.M., Senthil-Kumar M. and Mysore K.S. Agroinoculation and agroinfiltration: simple tools for complex gene function analyses. In: Pereira, A. (Ed.), Plant Reverse Genetics. Humana Press, 2011; 65-76.
CrossRef - Wieczorek P., Budziszewska M., Frąckowiak P. and Obrępalska-Stęplowska A., 2020. Development of a New Tomato Torrado Virus-Based Vector Tagged with GFP for Monitoring Virus Movement in Plants. Viruses 2020; 1195.
CrossRef - Tu L., Wu S., Gao D., Liu Y., Zhu Y. and Ji Y. Synthesis and Characterization of a Full-Length Infectious cDNA Clone of Tomato Mottle Mosaic Virus. Viruses. 2021; 1050.
CrossRef - Prema G.U. and Rangaswamy K.T. Molecular detection and characterization of coat protein gene of Mungbean Yellow Mosaic Virus (MYMV) from Karnataka. J. Agril. Sci. 2018; 5118-5122.
CrossRef - Sambrook J. and Russel D.W. Extraction and purification of plasmid; screening of bacterial colonies by hybridization. Molecular Cloning: a laboratory manual, 3rd Ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA. 2001
CrossRef - Dean F.B., Nelson J.R., Giesler T.L. and Lasken R.S. Rapid amplification of plasmid and phage DNA using phi29 DNA polymerase and multiply-primed rolling circle amplification. Res, 2001; 1095-1099.
CrossRef - Usharani K.S., Periasamy, M and Malathi V.G. Studies on the activity of a bidirectional promoter of Mungbean yellow mosaic India virus by agroinfiltration. Virus research, 2006; 154-162.
CrossRef - Biswas K.K and Varma A. Evaluation of resistance in blackgram (Phaseolus mungo) to variants of mungbean yellow mosaic geminivirus. Indian J. Agril. Sci. 2001; 215-218.
- Mandal B., Varma A. and Malathi V.G. Systemic infection of Vigna mungo using the cloned DNAs of the blackgram isolate of mungbean yellow mosaic geminivirus through agroinoculation and transmission of the progeny virus by whiteflies. Phytopathology. 1997. 145; 505-510.
CrossRef - Kumar S., Tanti B., Mukherjee S.K and Sahoo L. Molecular characterization and infectivity of Mungbean Yellow Mosaic India virus associated with yellow mosaic disease of cowpea and mungbean. Biocat Agri Biotech 11, 183–191.
CrossRef - Jyothsna P., Rawat R and Malathi V.G. Molecular characterization of a new begomovirus infecting a leguminous weed Rhynchosia minima in India. Virus Genes, 2011; 407-414.
CrossRef - Haq Q.M.I., Rouhibakhsh A., Ali A and Malathi V.G. Infectivity analysis of a blackgram isolate of Mungbean yellow mosaic virus and genetic assortment with MYMIV in selective hosts. Virus Genes, 2011; 429-439.
CrossRef - Briddon R.W and Markham P.G. Use of PCR in the detection and characterization of geminiviruses. OEPP, 1995; 315-320.
CrossRef - Biswas K.K., Tarafdar A. and Biswas K. Viral diseases and its mixed infection in mungbean and urdbean: major biotic constraints in production of food pulses in India. Modern Trends in Microbial Biodiversity of Natural Ecosystem. 2012; 301-317.
- Kassanis B., The transmission of sugar‐beet yellows virus by mechanical inoculation. Annals of Applied Biology, 1949; 270-272.
CrossRef - Saad M.F.M., Sau A.R., Akbar M.A., Baharum S.N., Ramzi A.B., Talip N and Bunawan H. Construction of Infectious Clones of Begomoviruses: Strategies, Techniques and Applications. Biology, 2021; 604.
CrossRef - Singh S., Nirmalkar V.K and Awasthi L.P. Recent advances in begomovirus research in India. Applied Plant Virology. 2020; 493-513.
CrossRef - Sacristan S., Diaz M., Fraile A and García-Arenal F. Contact transmission of Tobacco mosaic virus: a quantitative analysis of parameters relevant for virus evolution. virology, 2011; 4974-4981.
CrossRef - Jalender P., Bhat B.N., Anitha K and Vijayalakshmi K. Studies on transmission of cucumber mosaic virus (CMV) by sap inoculation in tomato. J. Pure and App. Biosci. 2017; 1908-1912.
CrossRef - Sundaresha S., Sreevathsa R., Balol G.B., Keshavareddy G., Rangaswamy K.T. and Udayakumar M. A simple, novel and high efficiency sap inoculation method to screen for tobacco streak virus. Mol. Biol Plants. 2012; 365-369.
CrossRef - Givord L. and Hirth L. Identification, purification and some properties of a mosaic virus of okra (Hibiscus esculentus). Annals of Applied Biol. 1973; 359-370.
CrossRef - Gonsalves D. Control of papaya ringspot virus in papaya: a case study. Annual review of phytopathol. 1998; 415-437.
CrossRef - Surekha S., Magar S.J., Satyadev P and Prasanna K.V. Transmission Studies on an Indian Isolate of Cowpea Mosaic Virus. J. Curr. Microbiol. App. Sci. 2018; 528-534.
CrossRef - El-Abhar M.A. El-Abhar M.A. Elkady K.M. Ghanem H.A. Bosila. Identification, characterization and ultrastructure aspects of Alfalfa mosaic virus infecting potato (Solanum tuberosum L.) in Egypt. J. Virol. Sci. 2018; 68–77.
- Zhang F., Liu S., Zhang T., Ye Z., Han X., Zhong K., Yang J., Chen J. and Liu P. Construction and biological characterization of an infectious full-length cDNA clone of a Chinese isolate of Wheat yellow mosaic virus. Virology. 2021; 101-109.
CrossRef - Liu, Y., Schiff, M. and Dinesh‐Kumar, S.P., 2002. Virus‐induced gene silencing in tomato. The Plant Journal, 31(6), pp.777-786.
CrossRef - Liu E and Page J.E. Optimized cDNA libraries for virus-induced gene silencing (VIGS) using tobacco rattle virus. Plant methods, 2008; 1-13.
CrossRef - Lu R., Malcuit I., Moffett P., Ruiz M.T., Peart J., Wu A.J., Rathjen J.P., Bendahmane A., Day L. and Baulcombe D.C. High throughput virus‐induced gene silencing implicates heat shock protein 90 in plant disease resistance. The EMBO journal, 2003; 5690-5699.
CrossRef - Dong, Y., Burch-Smith, T.M., Liu, Y., Mamillapalli, P. and Dinesh-Kumar, S.P., 2007. A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and-2 in floral development. Plant physiology, 145(4), pp.1161-1170.
CrossRef - Anand A., Vaghchhipawala Z., Ryu C.M., Kang L., Wang K., del-Pozo O., Martin G.B. and Mysore K.S. Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced gene silencing. Molecular Plant-Microbe Interactions, 2007; 41-52.
CrossRef - Gao S., Qu J., Chua N.H. and Ye J. A new strain of Indian cassava mosaic virus causes a mosaic disease in the biodiesel crop Jatropha curcas. Archives of virology, 2010; 607-612.
CrossRef - Wieczorek P., Budziszewska M. and Obrępalska-Stęplowska A. Construction of infectious clones of tomato torrado virus and their delivery by agroinfiltration. Archives of virology, 2015; 517-521
CrossRef - Usharani K.S., Periasamy M. and Malathi V.G. Studies on the activity of a bidirectional promoter of Mungbean yellow mosaic India virus by agroinfiltration. Virus research, 2006; 154-162.
CrossRef - Jacob SS, Vanitharani R, Karthikeyan AS, Chinchore Y, Thillaichidambaram P, Veluthambi K. Mungbean yellow mosaic virus-Vi Agroinfection by codelivery of DNA A and DNA B from one Agrobacterium strain. Plant Dis 2003; 247–251.
CrossRef - Sudha M., Karthikeyan A., Nagarajan P., Raveendran M., Senthil N., Pandiyan M., Angappan K., Ramalingam J., Bharathi M., Rabindran R. and Veluthambi K. Screening of mungbean (Vigna radiata) germplasm for resistance to Mungbean yellow mosaic virus using agroinoculation. Canadian J plant pathol. 2013; 424-430.
CrossRef - Kil E.J., Byun S., Kim S., Kim J., Park J., Cho S., Yang D.C., Lee K.Y., Choi H.S., Kim J.K. and Lee S. Sweet pepper confirmed as a reservoir host for tomato yellow leaf curl virus by both agro-inoculation and whitefly-mediated inoculation. Archives of virology, 2014; 2387-2395.
CrossRef - Koeda S., Homma K., Tanaka Y., Onizaki D., Kesumawati E., Zakaria S. and Kanzaki S., Inoculation of capsicums with Pepper yellow leaf curl Indonesia virus by combining agroinoculation and grafting. The Horticulture Journal, 2018; 364-371.
CrossRef - Kheyr‐Pour A., Gronenborn B. and Czosner H. Agroinocu.lation of tomato yellow leaf curl virus (TYLCV) overcomes the virus resistance of wild Lycopersicon Plant Breeding. 1994;228-233.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





