How to Cite | Publication History | PlumX Article Matrix
K. Sai Pavithra1* , Jeyanthi Annadurai1
, Jeyanthi Annadurai1 , K. Ramanathan2
, K. Ramanathan2 and R. Ragunathan3
and R. Ragunathan3
1Department of Biotechnology, Thanthai Hans Roever College of Arts and Science, Affiliated to Bharathidasan University, Perambalur, India.
2Department of Microbiology, Thanthai Hans Roever College of Arts and Science, Affiliated to Bharathidasan University, Perambalur, India.
3Centre for Bioscience and Nanoscience Research, Affiliated to Bharathiar University, Coimbatore, India.
Corresponding Author E-mail: penguinpavi@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3004
ABSTRACT:
Objectives: To identify the potential targets which effectively bind with the Interleukin Receptor associated tyrosine kinase. Methods: We analysed the potential targets to be docked using AutoDock V4.2 software. The database used to retrieve the structure is Protein Data Bank (PDB) on (12/02/2021). The docking parameters such as binding affinity and binding energy was determined. The core docking score was predicted when docking was performed and the results obtained from the binding parameters were compared with the docking parameters results. From the Gas Chromatography-Mass Spectrometry results, we found the potential targets to be docked with the Interleukin Receptor assisted tyrosine kinase. Findings: The targets 2-Propenoic acid, 3-phenyl-, ethyl ester, and hexadecanoic acid effectively docked with the receptor and predicts the binding energy (-5.83) and (-4.80). We are investigating the Artemisia pallens which is having good antidiabetic activity and based on the references, we found that Interleukin Receptor Associated Kinase is responsible for diabetes. Novelty/Improvement: From these investigations, we concluded that hexadecanoic acid, sourced from Artemisia pallens, successively binds with the receptor and suggests that it may be viable to discover drugs in the future.
KEYWORDS: Artemisia pallens; Diabetes; hexadecanoic acid; Interleukin Receptor Associated Kinase; 2-Propenoic acid; 3-phenyl- ethyl ester
Download this article as:| Copy the following to cite this article: Pavithra K. S, Annadurai J, Ramanathan K, Ragunathan R. Molecular Docking Approach for Predicting the Binding Affinity of Potential Targets with Interleukin Receptor Associated Kinase with Phytochemicals from Artemisia Pallens. Biosci Biotech Res Asia 2022;19(2). |
| Copy the following to cite this URL: Pavithra K. S, Annadurai J, Ramanathan K, Ragunathan R. Molecular Docking Approach for Predicting the Binding Affinity of Potential Targets with Interleukin Receptor Associated Kinase with Phytochemicals from Artemisia Pallens. Biosci Biotech Res Asia 2022;19(2). Available from: https://bit.ly/3aSCjYC |
Introduction
The (Interleukin Receptor Associated Kinase) IRAK-1, IRAK-2, IRAK-3 and IRAK-4 constitutes the serine threonine kinases which help in mediating the toll like receptors and the interleukin (IL-1) signalling pathways. (Jack et al., 2018) It is utilized in molecular docking because IRAK performs a key role in drug designing purpose to create an appropriate antidiabetic drug. IRAK is stated to play an important role in diabetes. It reduces the blood glucose level and improves the insulin level thereby acts as a good candidate for suitable diabetic medication.
Phosphorylation of proteins is catalysed by the protein kinases. Here, the phosphate groups are transferred from the phosphate donating molecule to the specific substrate. As the general structure and characteristics of kinase domains are prominent, protein kinases are the suitable candidates in the structure based drug discovery. (Myeong et al., 2018)
The Artemisia pallens Wall. ex DC is a xerophytic plant of the Asteraceae family. This aromatic herb is prevalent in Southern India. This plant is commercially used in treating various ailments.(Honmore et al.,2021) This plant was considered for research purposes because detailed antidiabetes work has not been carried so far. This plant is easily available and is a tropical plant of India. Phytochemical analysis of the plant was carried out using water, methanol, chloroform, diethyl ether, and n-butanol. Since the molecular docking studies have not been explored in this plant I decided to do the molecular docking studies on Artemisia pallens.
Materials and Methods
Preparation of the Plant Extract
The experimental plants Artemisia pallens are procured from the flower vendors from local markets (Coimbatore, Tamilnadu). The samples are transported into the laboratory and washed which removes the dirt from the plants and then air dried in a shady place. Plant leaves are collected and ground well by using mortar with pestle. Powdered plant products are dissolved by using methanol.
Molecular docking
The phytochemical compounds are selected based on the GCMS results and it is subjected to docking for the identification of potential targets. The analysis of the essential oils was carried out by gas chromatography coupled with mass spectrometry (GM-MS). Coupling was performed and equipped with a fused silica capillary column. The retention index were determined by gas chromatography on two fused silica capillary column’s (25m x 0.25 mm) of the type ov-101 and cabowax 20m with temperature programming identical to that used for the coupling. The structure of the target receptor Interleukin Receptor Associated Kinase (IRAK) was retrieved from protein data bank and the structure of the targets 2-Propenoic acid, 3-phenyl-, ethyl ester were retrieved from pubchem database. The structures were docked and calculated the binding affinity. The binding energy and bond specifications are calculated for the docked structure. The lowest binding energy, mean binding energy and bond specifications were compared for the docked complexes. IRAK is determined and identified using kit method.
 |
Graph 1: The GCMS results of 2-Propenoic acid, 3-phenyl-, ethyl ester and Hexadecanoic acid has been provided. |
Results and Discussion
The structures were retrieved and docking was performed to calculate the binding parameters. The name of the receptor with PDB ID 2OIB was described in Table-1 the target 2-Propenoic acid, 3-phenyl-, ethyl ester with Pubchem ID 7649 was described in Table-2. Binding energies for the docked complexes were predicted at different runs which were represented in Table 3. It shows the lowest binding energy (-6.01) and mean binding energy (-5.83) for the docked complexes. The target binds with the receptor in the location (TYR262) with bond length (2.37 A) was represented in Table 4. It has highest binding affinity with the receptor and the docked complexes of 2-Propenoic acid, 3-phenyl-, ethyl ester with Interleukin Receptor Associated Kinase was shown in Fig-1 and this picture labels the amino acid residues with their location. The binding locations of the docked complexes were shown in Fig-2. The target Hexadecanoic acid with Pubchem ID 985 was depicted in Table-5. Binding energies for the docked complexes were predicted at different runs which were represented in Table 6. It shows the lowest binding energy (-4.80) and mean binding energy (-4.80) for the docked complexes. The target binds with the receptor in the location (LYS213) with bond length (2.84 A) was represented in Table-7. It shows the highest binding affinity with the receptor. The docked complexes of Hexadecanoic acid with Interleukin Receptor Associated Kinase and the amino acid residues with their location was shown in Fig-3. The binding locations of the docked complexes were represented in Fig-4 IRAK improves the insulin level and also assist in the maintenance of the blood sugar levels. Various studies related to diabetes and IRAK has only been carried out, but combined studies related to the four aspects like IRAK, molecular docking, herbal medicine and diabetes has not been performed. So it may be considered that this study can be unique and different.
Table 1: Description about Interleukin Receptor Associated Kinase
| S.No | Receptor Name | PDB Id |
| 1. | Interleukin Receptor Associated Kinase (IRAK) | 2OIB |
Table 2: Description about 2-Propenoic acid, 3-phenyl-, ethyl ester Ligand.
| S.No | Ligand Name | Pubchem Id |
| 1. | 2-Propenoic acid, 3-phenyl-, ethyl ester | 7649 |
Table 3: Binding energies at different runs for the docked complex.
| Cluster rank | Lowest binding energy | Run | Mean binding energy |
| 1 | -6.01 | 8 | -5.83 |
| 2 | -5.86 | 7 | -5.75 |
Table 4: Bond specifications for the docked complexes.
| S.No | Type of bond | Amino acid residue | Ligand atom | Bond length
(Å) |
| 1 | Hydrogen | TYR262 | O | 2.37 |
 |
Figure 1: Docked complex of 2-Propenoic acid –, ethyl ester with Interleukin Associated Receptor Kinase. |
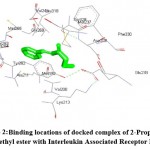 |
Figure 2: Binding locations of docked complex of 2-Propenoic acid –, ethyl ester with Interleukin Associated Receptor Kinase. |
Table 5: Description about Hexadecanoic acid.
| S.No | Ligand Name | Pubchem Id |
| 1. | Hexadecanoic acid | 985 |
Table 6: Binding energies at different runs for the docked complexes.
| Cluster rank | Lowest binding energy | Run | Mean binding energy |
| 1 | -4.80 | 2 | -4.80 |
| 2 | -4.22 | 1 | -4.20 |
Table 7: Bond specifications for the docked complexes
| S.No | Type of bond | Amino acid residue | Ligand atom | Bond length
(Å) |
| 1 | Hydrogen | LYS213 | O | 2.846845 |
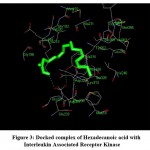 |
Figure 3: Docked complex of Hexadecanoic acid with Interleukin Associated Receptor Kinase |
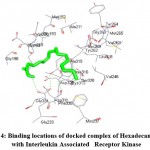 |
Figure 4: Binding locations of docked complex of Hexadecanoic acid with Interleukin Associated Receptor Kinase. |
Conclusion
The study identified the binding energy between the IRAK receptor and the hexadecanoic acid and the ligand 2-Propenoic acid, 3-phenyl-, ethyl ester. We observed that hexadecanoic acid effectively docked with the receptor indicating a good remedy for diabetes. The outcome of the results fulfils the objective of the study. From these investigations, we concluded that this medicinal herb can reduce the diabetic complications and it could be used in the process of drug discovery in future.
Acknowledgement
The author is thankful to the director and to the staff members of Centre for Bioscience and Nanoscience Research and also to the faculty members of the Department of Microbiology, Thanthai Hans Roever College for providing the necessary support to carry out this work.
Conflict of Interest
There is no conflict of interest.
Funding Sources
There is no funding source.
References
- Jack W. Singer, Angela Fleischman, Suliman Al-Fayoumi, John O. Mascarenhas, Qiang Yu, Anupriya Agarwal, Inhibition of interleukin-1 receptor-associated kinase 1 (IRAK1) as a therapeutic strategy, Oncotarget (2018), Vol 9, Issue 70, 33416-33439, doi: 10.18632/oncotarget.26058.
CrossRef - Myeong Hwi Lee, Anand Balupuri, Ye-rim Jung, Sungwook Choi, Areum Lee Young Sik Cho, Nam Sook Kang, Design of a Novel and Selective IRAK4 Inhibitor Using Topological Water Network Analysis and Molecular Modeling Approaches, Molecules, (2018), 29;23(12):3136, doi: 10.3390/molecules23123136.
CrossRef - Honmore, V. S., Natu, A. D., Khedkar, V. M., Arkile, M. A., Sarkar, D., & Rojatkar, S. R. (2021). Two antibacterial spiro compounds from the roots of Artemisia pallens wall: evidence from molecular docking. Natural Product Research, 1–8. doi:10.1080/14786419.2021.1902325
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





