How to Cite | Publication History | PlumX Article Matrix
Jehan Alrahimi 1,2 , Alia Aldahlawi1,2
, Alia Aldahlawi1,2 , Shahira Hassoubah1
, Shahira Hassoubah1 , Saeedah Al-Jadani 3
, Saeedah Al-Jadani 3 , Walaa Alyamani 1
, Walaa Alyamani 1 and Najla Alotaibi 1,2*
and Najla Alotaibi 1,2*
1Department of Biological Sciences, Faculty of Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
2Immunology Unit, King Fahad Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia
3Department of Basic Sciences, Faculty of Applied Medical Sciences, Al-Baha University, Al-Baha, Saudi Arabia.
Corresponding Author E-mail: najlabio@hotmail.com
DOI : http://dx.doi.org/10.13005/bbra/3010
ABSTRACT: Legionnaires’ disease (LD) is a type of severe pneumonia that mainly caused by bacteria of the genus Legionella. LD bacteria reside in the water systems of facilities where lack of water exchange or flow plays a crucial role in enhancing bacterial growth. The under-recognition of the dangers of Legionella along with easing of Coronavirus disease 2019 (COVID-19) lockdown restrictions and global reopening, pose a potential increased risk of developing LD. Various Legionella species can lead to legionellosis infections, including LD and Pontiac fever. Legionellosis cases is generally found in natural or artificial aquatic environments such as cooling towers, hot water tanks, or air conditioning. The bacteria elude the host’s immune responses by various strategies, including releasing effector proteins. Thus, this review provides insight into the microbiology, epidemiology, and host cell biology of L. pneumophila, as well as an emphasis on the bacterial novel survival strategies of L. pneumophila. Also, suggests taking intensive actions towards closed buildings as a potential source of bacterial infection.
KEYWORDS: COVID-19 restrictions; Epidemiology;Legionnaires' disease; Legionella pneumophila; T4SS, LCV
Download this article as:| Copy the following to cite this article: Alrahimi J, Aldahlawi A, Hassoubah S, Al-Jadani S, Alyamani W, Alotaibi N. An Insight into the Microbiology, Epidemiology, and Host Cell Biology of Legionella Pneumophila: A Review of Literature. Biosci Biotech Res Asia 2022;19(3). |
| Copy the following to cite this URL: Alrahimi J, Aldahlawi A, Hassoubah S, Al-Jadani S, Alyamani W, Alotaibi N. An Insight into the Microbiology, Epidemiology, and Host Cell Biology of Legionella Pneumophila: A Review of Literature. Biosci Biotech Res Asia 2022;19(3). Available from: https://bit.ly/3DR1183 |
L. pneumophila is an aerobic gram-negative, flagellated, rod-shaped, intracellular waterborne bacterium of the Legionella genus and the causative agent of most LD cases (see Figure 1). Legionella was the first defined bacterium that intracellularly multiplied within protozoan (initially aquatic amoebae), that help in understanding the bacteria’s capacity to infect protozoa. This bacterium is ubiquitous, usually found in moist soil and water, freshwater systems are the main reservoir of L. pneumophila [1]. Although freshwater systems colonised by Legionella can disperse aerosols through showers, whirlpools, fountains, and cooling towers, L. pneumophila prefers to grow in hot water systems, including hot water tanks and hot tubes. Legionella can proliferate in many types of niches. It can live in planktonic form, co-existing mainly within multi-organismal biofilms or replicating inside an amoebae within a freshwater system. This commonly leads to influenza-like outbreaks caused by different Legionella species [2]. The inhalation of Legionella micro-aspiration results in both LD and a mild respiratory illness called Pontiac fever [3]. LD manifests as a pneumonia illness with a case fatality of almost 10%. Non-fatal Pontiac fever is a less severe flu-like illness caused by L. pneumophila, with symptoms of fever, chills, and headache [4]. Those most at risk of LD include the elderly, smokers, patients with chronic lung diseases (such as emphysema), and immunocompromised individuals (e.g. people with cancer or kidney failure) [5].
Since the disease was first identified in 1976, the highest number of LD outbreaks has been recorded in 2018, with approximately 10,000 cases in the US according to the CDC, different outbreaks have also been reported in such global locations as Canada, the United Kingdom, Italy, Sweden, Portugal, and Japan [6-9]. In general, the bacteria can cause both population and nosocomial pneumonia, with sporadic cases accounting for 85 %. Legionella species are responsible for up to 50% of cases of hospital-acquired pneumonia [10]. Recently, COVID-19 lockdown restrictions have raised concerns about water stagnation and the consequent facilitation of bacterial growth [11, 12]. Thus, controlling microbial infections through water quality monitoring in closed buildings is recommended. This article will review the literature associated with the study of L. pneumophila, including its microbiological and epidemiological features. The article will also briefly describe the host biology and pathogenicity mechanisms of the bacteria.
L. pneumophila
In 1976, more than 200 people developed mysterious severe pneumonia illnesses resulting in 34 deaths [1,13]. Since this outbreak’s causative agent was unknown, the US Centers for Disease Control and Prevention (CDC) investigated the source of the infection. Although the source of Legionella was then unknown, investigators hypothesised that the air conditioning of the hotel in which the patients had stayed was the source of the infection. The CDC and the US National Institutes of Health (NIH) were the first medical organisations to start researching LD, with CDC beginning one of their largest investigations to follow the source of the outbreak. This epidemiological investigation aimed to track the etiological agent using various laboratory techniques. In 1977, the microbiologist Joseph McDade discovered L. pneumophila as the etiological agent of the LD outbreak [14]. He described it as a rod-shaped, Gram-negative bacterium and named it after the members of the American Legion first affected with the illness [15, 16]. Two years later, another outbreak occurred in which investigators found that a hospital’s air conditioning cooling tower was the source of L. pneumophila [17, 18].
L. pneumophila is a facultative intracellular pathogen. It replicates within human alveolar macrophages to avoid phagolysosome fusion and maintain replication within the host cell [19]. The intracellular vesicle of the bacteria thus becomes vigorously motile and overwhelmed by the infection. Consequently, lysis of the host cell releases the bacterial progeny from the macrophage to the surrounding environment [19]. This increases the patient’s susceptibility to acute lung inflammation, particularly in immunocompromised people and the elderly. Following inhalation of the aerosols, L. pneumophila avoids degradation and controls the immune system to form Legionella Containing Vacuoles (LCV). The LCV then employ rough endoplasmic reticulum and mitochondria to support L. pneumophila intracellular replication. Additionally, L. pneumophila has developed mechanisms such as hijacking host cell functions by secreting effector proteins using unique secretion systems [20]. Effector proteins of L. pneumophila have exceeded 300 effectors. These proteins facilitate bacterial survival primarily through the acquisition of the host’s nutrients. The excessive activity of effector proteins, paradoxically, increases pro-inflammatory cytokines [21, 22]. In addition to effectors, virulence factors including flagella, type IV, and LPS play a role in enhancing L. pneumophila pathogenesis [23, 24].
Different species of Legionella have been associated with both community-acquired pneumonia and nosocomial pneumonia [25]. Furthermore, Legionella is generally motile and requires specific environmental conditions to grow, including the presence of cystine and iron. L. pneumophila replicates intracellularly within eukaryotic host cells such as protozoa and macrophages [24, 26]. The bacteria replicate in the human lung, where alveolar macrophages lead to phagocytosis of the bacteria after the inhalation of contaminated water aerosols [27]. Alveolar macrophages are thus considered the primary type of human cell associated with this infection. Moreover, the ability of L. pneumophila to adapt to different hosts and to infect humans is due to its high-volume acquisition of effector proteins and genes [20]. The genus Legionella have surpassed 60 species and more than 70 serogroups [28], and the numbers continue to increase [29]. Thirty serogroups have been successfully isolated from patients and associated with human disease [30]. Whereas L. pneumophila was found to be responsible for most LD cases (approximately 95%) compared to other Legionella species. The strain associated with nearly 84% of LD cases is serogroup one, found in natural habitats, followed by L. longbeachae (3.9%) [31]. These strains’ high virulence is due to various ecological and physiological features, such as the O-antigen proteins identified in serogroup one [32]. In addition, serogroups four and six are also associated with the disease [28, 33].
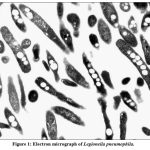 |
Figure 1: Electron micrograph of Legionella pneumophila. |
A morphology of virulent L. pneumophila cell with multiple intracellular inclusions. (Magnification was X 9,000).
Mode of transmission
Despite the severe outcomes of this disease, there is limited evidence of human-to-human transmission of L. pneumophila [34]. Consequently, this pathogen is known as ‘an accidental pathogen’ for which humans are the last host meaning that there is no subsequent transmission [27]. One mode of transmission for L. pneumophila to humans is through the inhalation of contaminated water droplets. The bacteria can reside and grow in artificial water systems, such as pipes, to form a biofilm [35]. Thus, L. pneumophila can cause disease only if it is inhaled or aspirated [36]. Serogroup one of L. pneumophila has been found across the United States, in 47% of cold-water of the publicly-used taps [37], followed by multiple LD’s outbreaks that have been related to several sources including contaminated cooling towers [38], closed-water distribution systems, and public whirlpool spas [39]. Other mechanisms and settings include hospital equipment, air conditioning, hotels, and cruise ships.
LD cases have also found to be associated with supermarket mist machines, fountains, and ice machines [40]. As the wide range of these settings makes clear, any aerosol generation source can transmit Legionella. Although Legionella antibodies have been found in animal sera, zoonotic transmission has not yet been detected [41]. However, co-infection may arise, particularly in immunocompromised patients [42]. Additionally, micropinocytosis plays a critical role in L. pneumophila pathogenicity; this process produces macropinosome, a vesicle generated from fusion of the membrane projections [43, 44]. It has been found that phospholipids such as phosphatidylinositol-3-kinase (PI3K) are involved in macropinosome formation [45]. Although the entry mechanisms of such protein’s remain unclear, it is important to mention that various structural genes, including RtxA and enhC, have a crucial role in facilitating pathogen transmission and attachment to host cells [46]. For instance, protein-protein interaction is facilitated by Sel1-like repeat (SLR), which is encoded by enhC [47]. Furthermore, Ca2+ binding is mediated by RtxA, which produces a total of eight motifs [48, 49].
Metabolic pathway
L. pneumophila replicates in both free-living amoebae and a host’s respiratory tract macrophages within LCV [50]. The formation of LCV, which are endoplasmic reticulum (ER)-associated compartments, involves a complex process [51]. That requires the bacteria to employ more than 300 effector proteins, including Defective Organelle Trafficking/Intracellular Multiplication (Dot/Icm) and to be translocated into the host’s cell by T4SS [52]. The wide variety of free-living protozoa explains why that L. pneumophila the most significant number of effector proteins compared to other bacteria. During replication, the membrane-bound compartment LCV protects the bacteria by preventing lysosomal degradation [53]. Also, within different ecological niches, the survival of L. pneumophila is attributed to the ability of LCV to facilitate the uptake of nutrients in the infected host cells [51]. In free-living protozoa, where the amino acids are the preferred carbon source for L. pneumophila [54].
L. pneumophila within the host cell can employ amino acid transporters to uptake the host amino acids as sources of carbon and energy. The effectors that L. pneumophila utilises to enhance the host’s amino acid acquisition and inhibit host translation of the proteins include Lgt1-3, SidI, SidL, LegK4, and RavX [55]. Although the role of translation elongation that resulted out of SidI binding to eEF1A and eEF1Bγ is poorly understood, this binding is not fully sufficient for impairing the translation [56]. While the mechanisms of RavX, SidI, and SidL remain unclear and require further investigations, it is known that the host’s polypeptide elongation process is inhibited by Lgt1-3 [57]. LegK4 can further induce phosphorylation of the host’s Hsp90 by reducing the host’s polypeptide refolding [58]. Furthermore, L. pneumophila highly up-regulates the gene synthesis of amino acids, leading to bacterial intracellular growth [59]. LCV-associated bacterial factors play a crucial role in the metabolic pathway of L. pneumophila. Additionally, Dot/Icm T4SS and Lsp type II secretion systems (T2SS) are essential for both intracellular and extracellular metabolism [50, 60]. The secreting effectors of T2SS plays a crucial role in L. pneumophila infection; more than 25 effector proteins are translocated by T2SS [61, 62]. This system has been associated with LCV membrane in host cytosol [63]. Thus, T2SS enhancing bacterial persistence in human lungs indicates its role in pathogenesis [64].
Epidemiological features of L. pneumophila
LD is considered a significant disease, and various countries including the US, Australia, Singapore, Canada, and New Zealand have developed LD surveillance schemes [65]. Nevertheless, globally reported LD data remain rare, contributing to under-recognition, lack of surveillance systems and diagnosis approaches. Resulting in limited data of LD incidence and other related disease-frequency measures [18]. Globally, case distributions are similar regarding both age and sex among countries. It has been shown that the disease is most common among elderly men, while it is uncommon among children [66]. The exact global incidence of LD is still unknown due to the lack of occurrence rates for detecting the disease. However, the US data shows an increase in LD crude incidence in the 21st century. Between 2000 and 2009, the incidence rate has increased from 3.9 per million to 11.5 per million from [67]. This data indicated a seasonal variation, in which approximately 63% of cases occurred in the summer and fall seasons. Incidence was also associated with travel history; almost 25% of patients contracted the illness while travelling [68]. According to the CDC, nearly 10,000 cases of LD were reported by US health departments in 2018 (CDC, 2018). A recent study in 2021, estimated that the LD cases true number is potentially two to three times higher than what is reported [69]. In consideration of the number of travel-associated cases, including those involving, hotel accommodations and cruise ships, effective disease surveillance systems have been created to collect, monitor, and manage data to assess public health actions by identifying sources and trends of infection [70].
Clinical outcomes
LD is atypical pneumonia; it may cause life-threatening respiratory disease, with severe to fatal infection in some cases [71, 72]. Clinically, LD may resemble pneumococcal pneumonia [73]. Although some studies have indicated a distinct clinical syndrome [74], others suggested that LD and pneumococcal pneumonia share the same clinical and radiographic presentation [75, 76]. Extrapulmonary and pneumonic complications including gastrointestinal and neurological signs are common in patients with community-acquired LD [77]. Symptomatic infection may occur outside the lung due to bacteraemia. The two manifestations of L. pneumophila are LD and Pontiac fever. The severity of LD ranges from mild to severe, and more severe pneumonia may require hospital admission [78]. LD has an incubation period between 7 and 14 days, symptoms begin 3 to 14 days after being the exposure. Symptoms include headache, shortness of breath, myalgia, cough, asthenia, and diarrhea [79].
Pontiac fever is characterised by a shorter incubation period than LD; in many cases, it develops within two days. The illness is further considered a self-limited disease and can be asymptomatic. A recent review summarised 136 outbreaks of LD and Pontiac fever between 2006 and 2017. With over 3,500 total cases, 115 outbreaks were LD, while only 4 were Pontiac fever. 17 outbreaks were mixed LD and Pontiac fever [80]. However, interpretation of Pontiac fever is limited due to the lacking an agreed-upon case definition by the scientific scholarly community for either probable or confirmed cases [80, 81]. The infection outcomes depend on bacterial virulence factors such as T4SS together with host immunity. Consequently, the elderly and individuals with chronic lung illnesses such as asthma are at higher risk of developing severe pneumonia [82, 83]. Severe hypoxemia and acute lung injury are also major clinical features of L. pneumophila induced pneumonia [84]. Furthermore, the serum of patients with L. pneumophila has shown high concentrations of inflammatory cytokines, including interferon-gamma (IFN-γ), tumour necrosis factor-alpha (TNFα), granulocyte-colony stimulating factor (GCSF), interleukin-12 (IL-12), IL-6, and IL-8, while IL-10 and IL-4 present with low or undetectable levels [85, 86].
Risk factors
Susceptibility to LD is associated with various host risk factors including smoking, advanced age, chronic cardiovascular, respiratory disease, receipt of a transplant, immune system compromise, diabetes, and alcohol abuse [82]. Also, at increased risk are malignant cancer and chemotherapy patients, including patients with hairy cell leukaemia [87], haematological malignancies [88], and solid tumours [89]. In addition, several reports have indicated infection in premature neonates and children [90]. Equally important are the risk factors related to the surrounding environment. Environmental risk factors associated with legionellosis outbreaks include travel, residency in particular facilities such as health care facilities, and poorly disinfected cooling towers [91]. Several recent studies have shown that LD follows seasonal patterns, with the most common activity in summer to early autumn [92]. These patterns are associated with environmental conditions including rainfall changes, climate, humidity, and temperature. Furthermore, nutrients are considered an essential ecological factor that facilitates the biofilm formation of L. pneumophila [93]. Many outbreaks have been connected with artificial environments that contain water at high temperatures. In particular, LD most often connected to air conditioning systems, cooling towers, and evaporative condensers [94]. Consequently, human-made aquatic reservoirs hold the potential to increase human susceptibility to Legionella, explaining the rapid increase in Legionella incidence in the latter half of the 20th century [95]. Additionally, incidence of the infection may increase during and after the COVID-19 pandemic [96, 97].
Antibiotic resistance of L. pneumophila
Antimicrobial resistance is a global challenge associated with morbidity and mortality. Although antibiotic resistance is unusual and not yet a major concern in treating L. pneumophila, it has been reported in several cases. A recent case of a patient with LD in the Netherlands presented an isolated fluoroquinolone (ciprofloxacin) resistance to L. pneumophila [98]. Also, antibiotic resistance in L. pneumophila has been identified in several countries such as China [99, 100]. In a study by Rahimi and Vesal, the highest resistance was against ciprofloxacin, erythromycin, clarithromycin, and moxifloxacin with resistance prevalence of 80%, 78%, 52%, and 48%, respectively. The lowest resistance was against rifampicin, doxycycline, and azithromycin with resistance prevalence of 19%, 22% and 26%, respectively [101]. Among macrolides antibiotics, clarithromycin shows high activity compared to azithromycin. In a recent study, minimum inhibitory concentrations were varied between L. pneumophila serogroup one and two, making levofloxacin more effective than either minocycline or doxycycline [102]. However, monotherapy involving erythromycin, ciprofloxacin, or
rifampicin is not recommended due to rapid antibiotic resistance [102, 103]. While erythromycin was the first choice for treating Legionella until the 1990s, it fell out of favour due to the side effects associated with intravenous delivery of the antibiotic. Furthermore, several epidemiological studies have shown that strains of L. pneumophila have high resistance against the most common antibiotics, including, ceftriaxone, clarithromycin, rifampicin, tigecycline, azithromycin, erythromycin, moxifloxacin, ciprofloxacin, and doxycycline [100-102]. Overall, regulatory treatment of L. pneumophila with levofloxacin and azithromycin has proven most effective in reducing transmission of L. pneumophila, and is recommended to treat both non- and immunocompromised individuals [104].
However, fluoroquinolones and macrolides achieve intracellular results therapeutic within tissue and particularly in macrophages, where the bacteria reside [105]. A low concentration of either macrolides or fluoroquinolones can inhibit various Legionella strains, [78]. Even though the prevalence of fluoroquinolone resistance may be underestimated, highlighting the importance of early Legionella infection diagnosis is crucial to ensure timely and accurate antibiotic treatment [106]. Digital PCR assay has proven helpful as a diagnostic tool to assess antibiotic therapy’s effectiveness [106]. Furthermore, PCR approach to detecting fluoroquinolone-resistant mutations of Legionella was implemented in 2017 [106]. Given the infrequency of recorded cases of resistance, the Infectious Diseases Society of America recommends either fluoroquinolones or macrolides as antimicrobial therapy [107]. In addition, a systematic review in 2021 have found no significant difference between fluoroquinolones and macrolides in term of effectiveness in decreasing mortality rate of patients with LD [107].
L. pneumophila increased risk during COVID-19
Limiting the growth of Legionella by first preventing L. pneumophila in building water systems is a potential preventive measure. If water is left in a system for more than a week without exchange or flow (e.g. water stagnation), the chance of bacterial growth will be increased [108]. The easing of COVID-19 lockdown restrictions and global reopening, along with under-recognition of the dangers of Legionella, pose a potential increased risk of developing LD [108]. Water temperature changes also provide a favourable environment for the bacteria to maintain growth. A recent study, a case of Legionella pneumonia is directly linked with a restaurant’s dishwasher shortly after the SARS-CoV-2 outbreak was brought under control. That emphasised the urgent need of thoroughly inspecting the water systems of different facilities before reopening following closure. Legionella infections are among the respiratory infections that have been diagnosed following lockdown due to the COVID-19 pandemic [109]. Patients have also been diagnosed with Legionella and COVID-19 co-infection, which can be lethal if left untreated. As a result, the emerging COVID-19 pandemic, warnings of co-infection with other respiratory pathogens are on the rise all over the world. Legionella thrives in poorly treated building water supplies, and outbreaks of LD have been recorded more often in hotels, long-term care centres, and hospitals. COVID-19 infections may increase co-infections risk of Legionella patients that associated with infections waves, posing a significant risk to high-risk COVID-19 patients following the pandemic’s peak and possibly raising disease incidence and mortality. Legionella cases and outbreaks are likely to be an increasingly important public health concern compared to the situation before the COVID-19 pandemic [109].
Immune responses to Legionella infection
The immune system has developed different defensive mechanisms against intracellular pathogens, including L. pneumophila. Entry of the bacteria will result in the inflammatory response and activation of immune cells, including macrophages, B lymphocytes, and sometimes natural killer (NK) cells. Consequently, the innate immune response inhibits bacterial growth, mainly in macrophages [110]. Released IFN-γ via macrophages activates NK and T cells, which increase macrophage resistance against infection. Other released cytokines, such as tumour necrosis factor-alpha (TNF-α), increase macrophage bactericidal activity and enhance resistance against the disease [111]. The regulated production of pro-inflammatory cytokines has helped to clear L. pneumonia infection in vivo through the innate immune system [112]. Also, an accumulation of immune cells during the inflammatory phase of L. pneumophila was observed, including monocytes, dendritic cells (DCs), neutrophils, and T cells [113]. Engaging the bacterial pathogen with the host led to a disruption of the host’s autoimmune defence mechanisms. In several cases, various cellular processes have been hijacked at the protein level by effector proteins, such as the hijacking of the host cytoplasmic, glycerol kinase enzyme by L. pneumophila to facilitate its metabolic process [114].
Moreover, effector proteins hijack different cellular functions to support bacterial intracellular replication. Accordingly, these effectors can bind, mimic, and modify the host’s proteins, including regulatory elements, enzymes, or transcription factors. Furthermore, the pathogen’s survival within the host cell depends on the formation of LCV to maintain replication [51]. LCV depends on effector proteins to enter the host cell and survive. Effectors alone have various structures, which raises questions about phenotypes relating to functions. Legionella inside the host cell can modulate the host signalling pathways through the secretion of effector proteins. The effectors are secreted through the secretion system, primarily a membrane complex called the Type IV secretion system (T4SS), which will be discussed in virulence factors section. There is also a correlation between the intensity of cytokine responses and patient severity [115]. The pro-inflammatory cytokines, such as TNFα in autoimmune patients, indicate susceptibility to acquiring LD [116]. Consequently, TNFα plays a crucial role in L. pneumophila induced pneumonia pathogenesis. In addition to s retrospective analysis study, it has shown that non-LD patients released a higher concentration of IFN-γ in response to bacterial lipopolysaccharides (LPS) than patients with LD. These results suggested that low IFN-γ levels may be associated with bacterial infection susceptibility [85]. Several in vitro studies supported Th-1 cytokines production via macrophage cells playing a role in restricting bacterial replication [117]. A strong inflammatory response is essential for limiting LD infection in the interaction between L. pneumophila and the adaptive immune response.
Conversely, T and B cells play a critical role in clearing existing infection [118]. T cells become activated after presentation of the bacterial antigens through antigen-presenting cells (APC). APC will uptake the antigen and process it into small peptides, then upload it on their major histocompatibility complex to identify T cell receptors (TCR). The most professional APC is DC which has been proven to initiate a specific immune response against L. pneumophila in mice [113]. Notably, in vitro study of bone marrow-derived dendritic cells (BMDCs) infected with L. pneumophila demonstrated that BMDCs induce the production of IFN-γ by CD4+ T cells. Simultaneously, the activation of CD4+ T cells is associated with LCV [110].
Virulence factors
Several studies have identified the virulence factors of L. pneumophila. They are associated with pathogenic strains, and are necessary to complete the intracellular infection cycle [24, 119] . Factors related to the L. pneumophila surface structure enhance pathogenesis and promote several processes – for instance, attachment to host cells and intracellular replication (see Table 1.). Those factors include an outer membrane protein (prion) [120], type IV pili [121], LPS, flagella, T2SS [60], and PilY1 protein [122].
Type IV secretion system
T4SS is considered major factor of the virulence factors of L. pneumophila; it is a complex protein nanomachine that bacteria utilise to promote proteins and DNA substrates into host cells [20]. The two phylogenetic types of T4SS are IVA and IVB [52]. The latter is represented by L. pneumophila Dot/Icm T4SS with over twenty proteins and encoded by 27 genes of the Dot/Icm. It includes essential proteins such as DotA, which plays a crucial role in T4SS assembly and activity [123, 124]. In addition, T4SS delivers more than 300 genetic and effector proteins of the bacteria to the host cell’s cytosol [125]. In a functional T4SS, bacteria can manipulate the trafficking of the host membrane, which allows them to escape phagolysosome fusion and facilitate bacterial replication. This can be implemented through remodelling the LCV into a rough ER-derived organelle [126]. Conversely, bacteria with a deficient stain of Dot/Icm T4SS, such as ΔdotA, cannot replicate intracellularly, because ΔdotA are fused with lysosomes and degraded after they traffic to the endocytic pathway [126-129].
T4SS effectors facilitate the intracellular replication of the bacteria by targeting the alveolar macrophages in the lung, injecting neutrophils, and harbouring live bacteria [130, 131]. Although L. pneumophila employs T4SS to inject effector proteins in macrophages, the type IV coupling complex (T4CC) is crucial for delivering the effector proteins to the T4SS [20]. Notably, the dot/icm DotL, including DotM and DotN, form T4CC [132]. Different types of effector proteins can be employed through T4CC binding sites. There are two main effector proteins, IcmS and IcmW, IcmSW dependent-effector and IcmSW independent-effector. The latter binds to the T4CC by DotL C-terminal sequence [20, 133].
Table 1: Virulence factors promoting L. pneumophila attachment.
|
VF |
Role |
References |
|
EnhC |
Promote intracellular growth by inhibiting the host’s innate immune response through reducing Nod1 and ensuring an efficient replication within macrophages through binding to L. pneumophila Slt.
|
[134] |
|
Lcl |
Facilitate invasion and cytokines expression. |
[135] |
|
Hsp60 |
Facilitate L. pneumophila entry, phagocytosis, and LCV development.
|
[24] |
|
type IV pili |
Facilitate adherence to host tissue, biofilm formation, and bacterial survival; promote horizontal gene transfer and enhance the bacterial adaptation to environment.
|
[52] |
|
LpnE |
Influence trafficking of the L. pneumophila vacuole.
|
[47, 136] |
|
RtxA |
Promote attachment and entry of host cells. |
[46] |
|
LadC |
Promote attachment to macrophages. |
[137] |
VF: Virulence factor.
PilY1
Pathogen attachment and entry are crucial to facilitating the pathogen’s modulation. L. pneumophila has various adherence determinants that enable entry into host cells, such as surface-associated hsp60 , type IV pilin gene [20], and the RtxA gene [46] (see Table 1.). One of the more recently described virulence factors of L. pneumophila is PilY1, which shares homology with other pathogens such as the PilY1 C-terminal domain of Pseudomonas aeruginosa and the PilC1/2 of Neisseria meningitidis, and Kingella kingae [138-140]. Accordingly, PilY1 is a cell surface protein that contributes to various virulence features, including biofilm formation and twitching motility [141]. A study conducted in 2017 showed that the deletion of PilY1 decreased the adhesion of both THP-1 macrophages and A549 epithelial cells to L. pneumophila. Simultaneously, reducing the replication rate in THP-1 macrophages, facilitate bacterial survival and replication [142].
Effector proteins and effector-triggered immunity
The initial process of recognition and elimination of pathogens results from the engagement between pathogen-associated molecular patterns (PAMPs) and the pathogen recognition receptors (PRRs) of the host. Toll-like receptors (TLRs) consider PRRs located on either plasma membrane or endosomal membrane playing an essential role in initiating an innate immune response against pathogens by recognising PAMPs [143]. As a result, pro-inflammatory cytokines are released to control the infection [111]. Nevertheless, bacterial pathogens have developed various virulence factors to avoid immune responses and increase their survival by acquiring the host’s nutrients [144]. The injection of the bacterial effectors into the host cell is done by highly-specialised secretion systems. Bacterial effectors are utilised by both intracellular and extracellular pathogens, highlighting the importance of these effector proteins in bacterial survival [145]. A process known as effector-triggered immunity (ETI) was first described in the immune response to plants’ pathogens [146]. ETI provides detection of the bacterial effector in many multicellular eukaryotes [145]. In plants, the ETI detects either the effectors or their intracellular activity, while in metazoans it detects only the intracellular effector activity [147]. In animals, the mechanism of the effectors is indirectly detected by cell-autonomous sensing of effectors’ homeostatic perturbations including pore formation [148].
All L. pneumophila strain encodes a special group of more than 300 effectors. Thus, the overall number of Legionella effectors to be investigated is override 300. The most studied strains are Philadelphia and Paris, each secreting roughly 330 effectors. Moreover, L. pneumophila has been described with more than 25 proteins released by T2SS and various secreted effectors by the Dot/Icm T4SS that exceed 300. One of the unique pathogenicity features of L. pneumophila is the total of 350 secreted proteins by L. pneumophila that does not correspond to any bacterial pathogen. This is due to the significant number of over 3,000 protein-coding genes per protein with a genome size of 3.2 Mb [149]. Furthermore, as a result of the protein-coding genes and the effector proteins, Coxiella burnetti is considered the closest bacterial pathogen to L. pneumophila. The pathogen has a genome size of 2Mb and more than 100 effector proteins, and 2,100 protein-coding genes [20].
Although effectors are essential for facilitating bacterial survival and growth, those effectors can paradoxically limit the bacteria by amplifying the production of the pro-inflammatory cytokines in macrophages. As a result, L. pneumophila is considered a useful pathogen model for understanding better effector-mediated immunity’s different mechanisms in detecting and eliminating the infection. The collective activity of the effectors leads to an increase in the inflammatory immune response against L. pneumophila [27]. For instance, Lgt1-3, SidI, and SidL effectors have been associated with activating IL-1α in macrophages [150]. Consequently, the selective upregulation of IL-1α results in an enhanced pro-inflammatory immune response and is considered significant in fighting L. pneumophila [112, 151]. The transitional inhibition which results from metabolic programming is facilitated by the effector-independent mechanism [21, 152].
Nevertheless, the host’s amino acid acquisition by L. pneumophila is due to effector-independent inhibition of host translation [153]. Transitional inhibition with pro-inflammatory response in the accidental host represents an example of a conical ETI [22]. Although some bacterial products, such as effectors, modulate unity, further studies are needed to investigate whether or not harnessing effectors can fight infectious diseases. One example of vaccine adjuvant, the TLR-9 agonist, CpG oligodeoxynucleotides (ODN), which has been used to amplify immune response against parasitic, bacterial, and viral pathogens, including the most recent SARS-CoV-2, the causative agent of COVID-19 [154, 155]. Furthermore, effectors that inhibit immunity have attracted attention as potential drug tools against inflammatory disease [156]. Since effectors require entry to the cytosol to be fully functional, the recent therapeutically uses of effectors have been implemented in 2017, through fusion to cell-penetrating peptides [157].
Detection and treatment
Microbial diseases are a real threat and considered one of the leading causes of death globally, particularly in developing countries, shedding light on the importance of accurate detection and identification of microbes. Time to detection is distinctly essential for LD patient outcomes, especially for at-risk individuals. Several detection methods for Legionella infection can be implemented via tissue, blood, or respiratory secretions such as sputum. Other methods using urine samples have also been established [158]. The most common methods for identifying the bacteria include microscopic and cultural techniques and serological tests [159]. Nevertheless, these more traditional methods have many limitations, encouraging the development of molecular strategies and the invention of Polymerase Chain Reaction (PCR) (Eklund, 2017). Biosensors are the cutting-edge technique in detecting microbial compounds such as proteins, enzymes, and DNA [77]. Furthermore, methods including urinary antigen tests and nucleic acid amplification testing have also been widely used [18]. Serology has been effective for historical epidemiological studies even where the infectious agent cannot be isolated despite clear evidence of LD [4]. However, one main limitation to serology is the false-positive occurrence that may result because of cross-reaction. As alternatives to serologic testing, the urinary antigen test and PCR-based detection methods are considered faster and more user-friendly.
As for pneumococcal pneumonia, the urinary antigen test has been used to detect the L. pneumophila serogroup one in particular has demonstrated a sensitivity of 70–100% and specificity of 95–100% [71]. The urinary antigen test also has the advantages of being inexpensive, straightforward, and rapid, making it a first-line screening tool [81, 160]. However, PCR, with the ability to detect a single pathogenic bacterium, is the most commonly used method. As a result of PCR sensitivity, false-positive results are less likely to occur compared to other methods. Additional advantages of PCR, include speed, high sensitivity, specificity, and accuracy owing to its ability to detect a small amount of nucleic acid [35]. Of course, L. pneumophila serogroup testing allows detection, however, improvements in assays identifying different serogroups and different Legionella species are required. PCR-based methods have become more commonly used in reference centres, such as L. pneumophila serogroup one detection centres. The development of a fast and accurate multiplexed real-time PCR assay can support other diagnostic methods [161]. Since L. pneumophila is considered a fastidious bacterium that grows and only slowly with complex nutrients, it is easily identified using biosensors. Such biosensors are a low-cost technique characterised by high specificity and sensitivity. A recent investigation of quantification biosensing of L. pneumophila, has shown that bioassay is an alternative conventional method for L. pneumophila detection [161].
Conclusion
Many water systems of closed buildings such as educational and business institutions, will have experienced water stagnation, providing a favourable environment for the growth of many bacteria including L. pneumophila. The intracellular L. pneumophila can exploit amoebae and also infect human macrophages. L. pneumophila is the causative agent of LD, a severe and potentially fatal form of pneumonia contracted by inhaling aerosols. L. pneumophila has developed complicated mechanisms to overcome environmental challenges and begin replicating within various niches, increasing its survival in the external environment. A complex regulatory network directs the shift between the two phases of non-virulent and virulent replication. This requires an engagement of both transcriptional and non-transcriptional regulatory elements to assure the effectiveness of the infection cycle. The metabolic changes trigger the morphological stress response, which results in nutrients availability in the surrounding environments. For example, the bacterial multiplication within LCV is supported by serine availability, which is used as a carbon and energy source and leads to increased metabolic activity. In addition, the stringent response of L. pneumophila that facilitates its survival under stress conditions in amino acid depletion. Under stress and starvation conditions, the bacteria enable the expression of virulence genes and shift the overall metabolism to use alternative carbon sources such as glucose. Thus, if these conditions last longer, L. pneumophila is ready to escape the host cell to start a new infection. There remains a serious lack of transmissive phase comprehensive analysis in vivo. Filling such gaps will provide insights into the usage of carbohydrates and crosstalk among the virulence regulatory elements. Urinary antigen tests and molecular methods are commonly used to diagnose infection. However, there are several advantages to effector-based therapeutic techniques in comparison to conventional biologics. Including high specificity, low concentration efficacy, cost-effectiveness, and autonomous translocation. To conclude, through investigation of water stagnation and an understanding of its role in the proliferation of Legionella is required along with lifting restrictions of COVID-19.
Acknowledgement
The authors would like to acknowledge the Mawakeb Alajer Association, Jeddah, Saudi Arabia through the Science Research and Innovation Unit at the Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia for facilitating in the conduction of this Research.
Conflict of Interest Statement
The authors declare no conflict of interest.
Funding source
This work was funded by Mawakeb Alajer Association, Jeddah, Saudi Arabia, through the Science Research and Innovation Unit at the Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia under research number (scigriu41/10).
References
- Kass EH. Legionnaires’ Disease. New England Journal of Medicine. 1977;297(22):1229-30.
- McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, Dowdle WR, et al. Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. New England Journal of Medicine. 1977;297(22):1197-203.
- McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, Dowdle WR. Legionnaires’ disease: isolation of a bacterium and demonstration of its role in other respiratory disease. The New England journal of medicine. 1977;297(22):1197-203.
- Brenner DJ, Steigerwalt AG, McDade JE. Classification of the Legionnaires’ disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann Intern Med. 1979;90(4):656-8.
- Dondero Jr TJ, Rendtorff RC, Mallison GF, Weeks RM, Levy JS, Wong EW, et al. An outbreak of Legionnaires’ disease associated with a contaminated air-conditioning cooling tower. New England Journal of Medicine. 1980;302(7):365-70.
- Faccini M, Russo AG, Bonini M, Tunesi S, Murtas R, Sandrini M, et al. Large community-acquired Legionnaires’ disease outbreak caused by Legionella pneumophila serogroup 1, Italy, July to August 2018. Eurosurveillance. 2020;25(20):1900523.
- Cadieux G, Brodeur J, Lamothe F, Lalancette C, Pilon PA, Kaiser D, et al. Community-based outbreak of Legionella pneumophila in Montréal Québec in 2019. CCDR. 2020;46(7/8).
- Almeida DQ, Silva T, Rodrigues V, Ladeira R, Sousa F, Capucho R, et al. Outbreak of Legionnaires’ Disease in the Northern Portuguese Coast During the COVID-19 Pandemic. Acta Médica Portuguesa. 2021;34(4):313-.
- Seto J, Amemura-Maekawa J, Sampei M, Araki K, Endo M, Kura F, et al. Investigation of a Legionnaires’ disease outbreak using direct sequence-based typing in Yamagata City, Japan, 2019. Japanese Journal of Infectious Diseases. 2021:JJID. 2020.815.
- Hashmi HRT, Saladi L, Petersen F, Khaja M, Diaz-Fuentes G. Legionnaires’ Disease: Clinicoradiological Comparison of Sporadic Versus Outbreak Cases. Clinical medicine insights Circulatory, respiratory and pulmonary medicine. 2017;11:1179548417711941.
- Nisar MA, Ross KE, Brown MH, Bentham R, Whiley H. Water stagnation and flow obstruction reduces the quality of potable water and increases the risk of legionelloses. Frontiers in Environmental Science. 2020.
- Nisar MA, Ross KE, Brown MH, Bentham R, Whiley H. Water Stagnation and Flow Obstruction Reduces the Quality of Potable Water and Increases the Risk of Legionelloses. Frontiers in Environmental Science. 2020;8(260).
- Van Heijnsbergen E, Schalk JA, Euser SM, Brandsema PS, den Boer JW, de Roda Husman AM. Confirmed and potential sources of Legionella reviewed. Environmental science & technology. 2015;49(8):4797-815.
- Jomehzadeh N, Moosavian M, Saki M, Rashno M. Legionella and legionnaires’ disease: An overview. Journal of Acute Disease. 2019;8(6):221.
- Hamilton K, Prussin A, Ahmed W, Haas C. Outbreaks of legionnaires’ disease and pontiac fever 2006–2017. Current environmental health reports. 2018;5(2):263-71.
- Wang C, Chuai X, Liang M. Legionella feeleii: pneumonia or Pontiac fever? Bacterial virulence traits and host immune response. Medical microbiology and immunology. 2019;208(1):25-32.
- Oggioni C, Za A, Auxilia F, Faccini M, Senatore S, Vismara C, et al. Legionnaires’ disease contracted from patient workplace: First report of a severe case of coinfection with varicella-zoster virus. American journal of infection control. 2016;44(10):1164-5.
- Graham FF, Hales S, White PS, Baker MG. Review Global seroprevalence of legionellosis-a systematic review and meta-analysis. Scientific reports. 2020;10(1):1-11.
- Escoll P, Rolando M, Gomez-Valero L, Buchrieser C. From amoeba to macrophages: exploring the molecular mechanisms of Legionella pneumophila infection in both hosts. Current topics in microbiology and immunology. 2013;376:1-34.
- Meir A, Macé K, Lukoyanova N, Chetrit D, Hospenthal MK, Redzej A, et al. Mechanism of effector capture and delivery by the type IV secretion system from Legionella pneumophila. Nature communications. 2020;11(1):1-11.
- Price C, Jones S, Mihelcic M, Santic M, Kwaik YA. Paradoxical pro-inflammatory responses by human macrophages to an amoebae host-adapted Legionella effector. Cell host & microbe. 2020;27(4):571-84. e7.
- Ngwaga T, Chauhan D, Shames SR. Mechanisms of Effector-Mediated Immunity Revealed by the Accidental Human Pathogen Legionella pneumophila. Frontiers in Cellular and Infection Microbiology. 2021;10(924).
- Liu X, Shin S. Viewing Legionella pneumophila pathogenesis through an Immunological Lens. Journal of molecular biology. 2019;431(21):4321-44.
- Chauhan D, Shames SR. Pathogenicity and Virulence of Legionella: Intracellular replication and host response. Virulence. 2021;12(1):1122-44.
- Miyashita N, Higa F, Aoki Y, Kikuchi T, Seki M, Tateda K, et al. Distribution of Legionella species and serogroups in patients with culture-confirmed Legionella pneumonia. Journal of Infection and Chemotherapy. 2020;26(5):411-7.
- Zayed AR, Pecellin M, Jaber L, Butmeh S, Bahader SA, Steinert M, et al. Cytotoxicity, Intracellular Replication, and Contact-Dependent Pore Formation of Genotyped Environmental Legionella pneumophila Isolates from Hospital Water Systems in the West Bank, Palestine. Pathogens. 2021;10(4):417.
- Ngwaga T, Chauhan D, Shames SR. Mechanisms of effector-mediated immunity revealed by the accidental human pathogen Legionella pneumophila. Frontiers in Cellular and Infection Microbiology. 2021;10:924.
- Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ disease: 25 years of investigation. Clinical microbiology reviews. 2002;15(3):506-26.
- Katsiaflaka A, Pournaras S, Kristo I, Mouchtouri VA, Kyritsi M, Velonakis E, et al. Epidemiological investigation of Legionella pneumophila serogroup 2 to 14 isolates from water samples by amplified fragment length polymorphism and sequence-based typing and detection of virulence traits. Applied and environmental microbiology. 2016;82(20):6102-8.
- Patrizia M, Annalisa B, Immacolata A, Isabella M, de Niederhäusern S, Moreno B. Protozoa and human macrophages infection by Legionella pneumophila environmental strains belonging to different serogroups. Archives of microbiology. 2013;195(2):89-96.
- Fliermans CB. Legionella Ecology. Bioaerosols: CRC Press; 2020. p. 49-76.
- Palusinska-Szysz M, Luchowski R, Gruszecki WI, Choma A, Szuster-Ciesielska A, Lück C, et al. The role of Legionella pneumophila serogroup 1 lipopolysaccharide in host-pathogen interaction. Frontiers in microbiology. 2019;10:2890.
- Paranjape K, Bédard É, Whyte LG, Ronholm J, Prévost M, Faucher SP. Presence of Legionella spp. in cooling towers: the role of microbial diversity, Pseudomonas, and continuous chlorine application. Water research. 2020;169:115252.
- Borges V, Nunes A, Sampaio DA, Vieira L, Machado J, Simões MJ, et al. Legionella pneumophila strain associated with the first evidence of person-to-person transmission of Legionnaires’ disease: a unique mosaic genetic backbone. Scientific reports. 2016;6(1):1-11.
- Hayatimehr S, Amirmozafari N, Masjedian F. Investigation of Virulence Genes and Biofilm Formation Among Legionella Pneumophila Isolated from Hospital Water Sources. 2020.
- Prussin AJ, Schwake DO, Marr LC. Ten questions concerning the aerosolization and transmission of Legionella in the built environment. Building and Environment. 2017;123:684-95.
- Donohue MJ, O’connell K, Vesper SJ, Mistry JH, King D, Kostich M, et al. Widespread molecular detection of Legionella pneumophila serogroup 1 in cold water taps across the United States. Environmental science & technology. 2014;48(6):3145-52.
- Young C, Smith D, Wafer T, Crook B. Rapid Testing and Interventions to Control Legionella Proliferation following a Legionnaires’ Disease Outbreak Associated with Cooling Towers. Microorganisms. 2021;9(3):615.
- Linsak DT, Kese D, Broznic D, Lusic DV, Cenov A, Moric M, et al. Sea water whirlpool spa as a source of Legionella infection. Journal of Water and Health. 2021;19(2):242-53.
- Alexander TY, Kamali A, Vugia DJ. Legionella epidemiologic and environmental risks. Current Epidemiology Reports. 2019;6(3):310-20.
- Pan X, Yang X. A serological investigation of Legionella infection in six-species of poultries and domestic animals in Luzhou City, Sichuan Province. Zhonghua liu xing bing xue za zhi= Zhonghua liuxingbingxue zazhi. 1999;20(2):108-10.
- Head BM, Trajtman A, Bernard K, Burdz T, Vélez L, Herrera M, et al. Legionella co-infection in HIV-associated pneumonia. Diagnostic microbiology and infectious disease. 2019;95(1):71-6.
- Cunha BA. Legionnaires’ disease: clinical differentiation from typical and other atypical pneumonias. Infectious disease clinics of North America. 2010;24(1):73.
- de Carvalho TMU, Barrias ES, de Souza W. Macropinocytosis: a pathway to protozoan infection. Frontiers in Physiology. 2015;6(106).
- Quinn SE, Huang L, Kerkvliet JG, Swanson JA, Smith S, Hoppe AD, et al. The structural dynamics of macropinosome formation and PI3-kinase-mediated sealing revealed by lattice light sheet microscopy. Nature Communications. 2021;12(1):1-12.
- Brown CL, Garner E, Jospin G, Coil DA, Schwake DO, Eisen JA, et al. Whole genome sequence analysis reveals the broad distribution of the RtxA type 1 secretion system and four novel putative type 1 secretion systems throughout the Legionella genus. PloS one. 2020;15(1):e0223033.
- Cirillo SLG, Lum J, Cirillo JD. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology (Reading, England). 2000;146 ( Pt 6):1345-59.
- Jacobi S, Heuner K. Description of a putative type I secretion system in Legionella pneumophila. International Journal of Medical Microbiology. 2003;293(5):349-58.
- Voth KA. Structural Biology of Legionella pneumophila Effectors: University of Saskatchewan; 2019.
- Manske C, Finsel I, Hoffmann C, Hilbi H. Analysis of Legionella Metabolism by Pathogen Vacuole Proteomics. Microbial Proteomics: Springer; 2018. p. 59-76.
- Steiner B, Weber S, Hilbi H. Formation of the Legionella-containing vacuole: phosphoinositide conversion, GTPase modulation and ER dynamics. International Journal of Medical Microbiology. 2018;308(1):49-57.
- Kim H, Kubori T, Yamazaki K, Kwak M-J, Park S-Y, Nagai H, et al. Structural basis for effector protein recognition by the Dot/Icm Type IVB coupling protein complex. Nature communications. 2020;11(1):1-11.
- Bärlocher K, Welin A, Hilbi H. Formation of the Legionella replicative compartment at the crossroads of retrograde trafficking. Frontiers in cellular and infection microbiology. 2017;7:482.
- Manske C, Hilbi H. Metabolism of the vacuolar pathogen Legionella and implications for virulence. Frontiers in cellular and infection microbiology. 2014;4:125.
- Belyi Y. Targeting Eukaryotic mRNA translation by Legionella pneumophila. Frontiers in molecular biosciences. 2020;7:80.
- Joseph AM, Pohl AE, Ball TJ, Abram TG, Johnson DK, Geisbrecht BV, et al. The Legionella pneumophila metaeffector Lpg2505 (MesI) regulates SidI-mediated translation inhibition and novel glycosyl hydrolase activity. Infection and immunity. 2020;88(5):e00853-19.
- De Leon JA, Qiu J, Nicolai CJ, Counihan JL, Barry KC, Xu L, et al. Positive and Negative Regulation of the Master Metabolic Regulator mTORC1 by Two Families of Legionella pneumophila Effectors. Cell Rep. 2017;21(8):2031-8.
- Moss SM, Taylor IR, Ruggero D, Gestwicki JE, Shokat KM, Mukherjee S. A Legionella pneumophila kinase phosphorylates the Hsp70 chaperone family to inhibit eukaryotic protein synthesis. Cell host & microbe. 2019;25(3):454-62. e6.
- Oliva G, Sahr T, Buchrieser C. The Life Cycle of L. pneumophila: Cellular Differentiation Is Linked to Virulence and Metabolism. Front Cell Infect Microbiol. 2018;8:3.
- White RC, Cianciotto NP. Assessing the impact, genomics and evolution of type II secretion across a large, medically important genus: the Legionella type II secretion paradigm. Microbial genomics. 2019;5(6).
- Ghosal D, Kim KW, Zheng H, Kaplan M, Truchan HK, Lopez AE, et al. In vivo structure of the Legionella type II secretion system by electron cryotomography. Nat Microbiol. 2019;4(12):2101-8.
- Portlock TJ, Tyson JY, Dantu SC, Rehman S, White RC, McIntire IE, et al. Structure, dynamics and cellular insight into novel substrates of the Legionella pneumophila type II secretion system. Frontiers in molecular biosciences. 2020;7:112.
- Truchan HK, Christman HD, White RC, Rutledge NS, Cianciotto NP. Type II Secretion Substrates of Legionella pneumophila Translocate Out of the Pathogen-Occupied Vacuole via a Semipermeable Membrane. mBio. 2017;8(3).
- White RC, Truchan HK, Zheng H, Tyson JY, Cianciotto NP. Type II secretion promotes bacterial growth within the Legionella-containing vacuole in infected amoebae. Infection and immunity. 2019;87(11):e00374-19.
- de Jong B, Hallström LP. European Surveillance of Legionnaires’ Disease. Current Issues in Molecular Biology. 2020;42(1):81-96.
- Foccillo G. The Infections Causing Acute Respiratory Failure in Elderly Patients. Ventilatory Support and Oxygen Therapy in Elder, Palliative and End-of-Life Care Patients: Springer; 2020. p. 35-45.
- Control CfD, Prevention. Legionellosis—United States, 2000-2009. MMWR Morbidity and mortality weekly report. 2011;60(32):1083-6.
- Garcia-Vidal C, Labori M, Viasus D, Simonetti A, Garcia-Somoza D, Dorca J, et al. Rainfall is a risk factor for sporadic cases of Legionella pneumophila pneumonia. PloS one. 2013;8(4):e61036.
- Collier SA, Deng L, Adam EA, Benedict KM, Beshearse EM, Blackstock AJ, et al. Estimate of burden and direct healthcare cost of infectious waterborne disease in the United States. Emerging infectious diseases. 2021;27(1):140.
- Papadakis A, Keramarou M, Chochlakis D, Sandalakis V, Mouchtouri VA, Psaroulaki A. Legionella spp. Colonization in Water Systems of Hotels Linked with Travel-Associated Legionnaires’ Disease. Water. 2021;13(16):2243.
- Schumacher A, Kocharian A, Koch A, Marx J. Fatal Case of Legionnaires’ Disease After Home Exposure to Legionella pneumophila Serogroup 3—Wisconsin, 2018. Morbidity and Mortality Weekly Report. 2020;69(8):207.
- Jiang L, Tao S, Mu D, Zhang N, Zhao L, Chen Y. Case report: fatal pneumonia caused by new sequence type Legionella pneumophilia serogroup 1. Medicine. 2020;99(43).
- Roed T, Schønheyder HC, Nielsen H. Predictors of positive or negative legionella urinary antigen test in community-acquired pneumonia. Infectious Diseases. 2015;47(7):484-90.
- Pierre DM, Baron J, Victor LY, Stout JE. Diagnostic testing for Legionnaires’ disease. Annals of clinical microbiology and antimicrobials. 2017;16(1):1-4.
- Cunha BA, Burillo A, Bouza E. Legionnaires’ disease. The Lancet. 2016;387(10016):376-85.
- Rech MM, Swalla BM, Dobranic JK. Evaluation of Legiolert for quantification of Legionella pneumophila from non-potable water. Current microbiology. 2018;75(10):1282-9.
- Kao W-F, Wang J-T, Sheng W-H, Chen Y-C. Community-acquired Legionnaires’ disease at a medical center in northern Taiwan. Journal of Microbiology, Immunology and Infection. 2019;52(3):465-70.
- Chahin A, Opal SM. Severe pneumonia caused by Legionella pneumophila: differential diagnosis and therapeutic considerations. Infectious Disease Clinics. 2017;31(1):111-21.
- Dalal N, Athwal PSS, Tharu B, Shah P, Shah L. Legionnaires Disease Presenting as Diarrhea: A Case Report. Cureus. 2020;12(9).
- Hamilton KA, Prussin AJ, Ahmed W, Haas CN. Outbreaks of Legionnaires’ Disease and Pontiac Fever 2006–2017. Current Environmental Health Reports. 2018;5(2):263-71.
- Ito A, Yamamoto Y, Ishii Y, Okazaki A, Ishiura Y, Kawagishi Y, et al. Evaluation of a novel urinary antigen test kit for diagnosing Legionella pneumonia. International Journal of Infectious Diseases. 2021;103:42-7.
- Cooley LA, Pondo T, Francois Watkins LK, Shah P, Schrag S, John ABCSPotEIPNVDANCMTSRPALEPED. Population-Based Assessment of Clinical Risk Factors for Legionnaires’ Disease. Clinical Infectious Diseases. 2020;70(11):2428-31.
- Hunter CM, Salandy SW, Smith JC, Edens C, Hubbard B. Racial Disparities in Incidence of Legionnaires’ Disease and Social Determinants of Health: A Narrative Review. Public Health Reports. 2021:00333549211026781.
- Kashif M, Patel R, Bajantri B, Diaz-Fuentes G. Legionella pneumonia associated with severe acute respiratory distress syndrome and diffuse alveolar hemorrhage-A rare association. Respiratory medicine case reports. 2017;21:7-11.
- Kak G, Raza M, Tiwari BK. Interferon-gamma (IFN-γ): exploring its implications in infectious diseases. Biomolecular concepts. 2018;9(1):64-79.
- Gomes TS, Gjiknuri J, Magnet A, Vaccaro L, Ollero D, Izquierdo F, et al. The Influence of Acanthamoeba–Legionella Interaction in the Virulence of Two Different Legionella Species. Frontiers in microbiology. 2018;9:2962.
- Radaelli F, Langer M, Chiorboli O, Proietti D, Baldini L. Severe legionella pneumophila infection in a patient with hairy cell leukemia in partial remission after a interferon treatment. Hematological oncology. 1991;9(3):125-8.
- Kugler JW, Armitage JO, Helms CM, Klassen LW, Goeken NE, Ahmann GB, et al. Nosocomial Legionnaires’ disease: occurrence in recipients of bone marrow transplants. The American journal of medicine. 1983;74(2):281-8.
- Marston BJ, Lipman HB, Breiman RF. Surveillance for Legionnaires’ disease: risk factors for morbidity and mortality. Archives of Internal Medicine. 1994;154(21):2417-22.
- Guyard C, Low DE. Legionella infections and travel associated legionellosis. Travel Medicine and Infectious Disease. 2011;9(4):176-86.
- Orkis LT, Harrison LH, Mertz KJ, Brooks MM, Bibby KJ, Stout JE. Environmental sources of community-acquired legionnaires’ disease: A review. International journal of hygiene and environmental health. 2018;221(5):764-74.
- Buchholz U, Altmann D, Brodhun B. Differential Seasonality of Legionnaires’ Disease by Exposure Category. International journal of environmental research and public health. 2020;17(9):3049.
- Abu Khweek A, Amer AO. Factors mediating environmental biofilm formation by Legionella pneumophila. Frontiers in cellular and infection microbiology. 2018;8:38.
- Hamilton KA, Hamilton MT, Johnson W, Jjemba P, Bukhari Z, LeChevallier M, et al. Health risks from exposure to Legionella in reclaimed water aerosols: Toilet flushing, spray irrigation, and cooling towers. Water research. 2018;134:261-79.
- Baron J, Morris L, Stout J. Control of Legionella in hospital potable water systems. Decontamination in Hospitals and Healthcare: Elsevier; 2020. p. 71-100.
- Dey R, Ashbolt NJ. Legionella Infection during and after the COVID-19 Pandemic. ACS ES&T Water. 2020;1(1):13-4.
- Verhasselt HL, Buer J, Dedy J, Ziegler R, Steinmann J, Herbstreit F, et al. COVID-19 Co-infection with Legionella pneumophila in 2 Tertiary-Care Hospitals, Germany. Emerging infectious diseases. 2021;27(5):1535.
- Bruin JP, Koshkolda T, EP IJ, Lück C, Diederen BM, Den Boer JW, et al. Isolation of ciprofloxacin-resistant Legionella pneumophila in a patient with severe pneumonia. The Journal of antimicrobial chemotherapy. 2014;69(10):2869-71.
- Jia X, Ren H, Nie X, Li Y, Li J, Qin T. Antibiotic resistance and azithromycin resistance mechanism of Legionella pneumophila serogroup 1 in China. Antimicrobial agents and chemotherapy. 2019;63(10):e00768-19.
- Pappa O, Chochlakis D, Sandalakis V, Dioli C, Psaroulaki A, Mavridou A. Antibiotic Resistance of Legionella pneumophila in Clinical and Water Isolates—A Systematic Review. International journal of environmental research and public health. 2020;17(16):5809.
- Rahimi B, Vesal A. Antimicrobial resistance properties of legionella pneumophila isolated from the cases of lower respiratory tract infections. Biomedical and Pharmacology Journal. 2017;10(1):59-65.
- Diaz-Fuentes G, Singhal R, Venkatram S. Advances in Treatment and Outcomes of Patients with Legionella Infection. Hospital Acquired Infection and Legionnaires’ Disease: IntechOpen; 2019.
- Nielsen K, Bangsborg JM, Høiby N. Susceptibility of Legionella species to five antibiotics and development of resistance by exposure to erythromycin, ciprofloxacin, and rifampicin. Diagnostic microbiology and infectious disease. 2000;36(1):43-8.
- Carratalà J, Garcia-Vidal C. An update on Legionella. Current opinion in infectious diseases. 2010;23(2):152-7.
- Jasper AS, Musuuza JS, Tischendorf JS, Stevens VW, Gamage SD, Osman F, et al. Are fluoroquinolones or macrolides better for treating Legionella pneumonia? A systematic review and meta-analysis. Clinical Infectious Diseases. 2021;72(11):1979-89.
- Hennebique A, Bidart M, Jarraud S, Beraud L, Schwebel C, Maurin M, et al. Digital PCR for Detection and Quantification of Fluoroquinolone Resistance in Legionella pneumophila. Antimicrobial agents and chemotherapy. 2017;61(9).
- Kato H, Hagihara M, Asai N, Shibata Y, Koizumi Y, Yamagishi Y, et al. Meta-analysis of fluoroquinolones versus macrolides for treatment of legionella pneumonia. Journal of Infection and Chemotherapy. 2021;27(3):424-33.
- Rhoads WJ, Hammes F. Growth of Legionella during COVID-19 lockdown stagnation. Environmental Science: Water Research & Technology. 2021;7(1):10-5.
- Palazzolo C, Maffongelli G, D’Abramo A, Lepore L, Mariano A, Vulcano A, et al. Legionella pneumonia: increased risk after COVID-19 lockdown? Italy, May to June 2020. Eurosurveillance. 2020;25(30):2001372.
- Horwitz MA. The immunobiology of Legionella pneumophila. Intracellular Parasitism: CRC Press; 2020. p. 141-56.
- Grigoryeva LS, Cianciotto NP. Human macrophages utilize a wide range of pathogen recognition receptors to recognize Legionella pneumophila, including Toll-Like Receptor 4 engaging Legionella lipopolysaccharide and the Toll-like Receptor 3 nucleic-acid sensor. PLoS Pathogens. 2021;17(7):e1009781.
- Liu X, Boyer MA, Holmgren AM, Shin S. Legionella-infected macrophages engage the alveolar epithelium to metabolically reprogram myeloid cells and promote antibacterial inflammation. Cell host & microbe. 2020;28(5):683-98. e6.
- Park B, Park G, Kim J, Lim SA, Lee K-M. Innate immunity against Legionella pneumophila during pulmonary infections in mice. Archives of pharmacal research. 2017;40(2):131-45.
- Häuslein I, Manske C, Goebel W, Eisenreich W, Hilbi H. Pathway analysis using 13C‐glycerol and other carbon tracers reveals a bipartite metabolism of Legionella pneumophila. Molecular microbiology. 2016;100(2):229-46.
- Endeman H, Meijvis S, Rijkers G, van Velzen–Blad H, Van Moorsel C, Grutters J, et al. Systemic cytokine response in patients with community-acquired pneumonia. European Respiratory Journal. 2011;37(6):1431-8.
- Bicheiro LMF. A systematic review on serious infections by the intracellular bacterial pathogens Legionella, Listeria, and Salmonella in patients receiving anti-TNF therapy. 2018.
- Horwitz MA. Influence of Interferon-Gamma on Legionella pneumophila: Mononuclear Phagocyte Interaction. Anti-Infective Applications of Interferon-Gamma: CRC Press; 2020. p. 265-78.
- Kikuchi T, Andarini S, Xin H, Gomi K, Tokue Y, Saijo Y, et al. Involvement of fractalkine/CX3CL1 expression by dendritic cells in the enhancement of host immunity against Legionella pneumophila. Infection and immunity. 2005;73(9):5350-7.
- Oliva G, Sahr T, Buchrieser C. The life cycle of L. pneumophila: cellular differentiation is linked to virulence and metabolism. Frontiers in cellular and infection microbiology. 2018;8:3.
- Younas F, Soltanmohammadi N, Knapp O, Benz R. The major outer membrane protein of Legionella pneumophila Lpg1974 shows pore-forming characteristics similar to the human mitochondrial outer membrane pore, hVDAC1. Biochimica et biophysica acta Biomembranes. 2018;1860(8):1544-53.
- Johnson DI. Legionella spp. Bacterial Pathogens and Their Virulence Factors: Springer; 2018. p. 279-87.
- Hoppe J, Ünal CM, Thiem S, Grimpe L, Goldmann T, Gaßler N, et al. PilY1 Promotes Legionella pneumophila Infection of Human Lung Tissue Explants and Contributes to Bacterial Adhesion, Host Cell Invasion, and Twitching Motility. Frontiers in Cellular and Infection Microbiology. 2017;7(63).
- Durie CL, Sheedlo MJ, Chung JM, Byrne BG, Su M, Knight T, et al. Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex. Elife. 2020;9:e59530.
- Kubori T, Nagai H. Isolation of the Dot/Icm type IV secretion system core complex from Legionella pneumophila. Legionella: Springer; 2019. p. 241-7.
- Kåhrström CT. Solving the T4SS structural mystery. Nature Reviews Microbiology. 2014;12(5):312-3.
- Ge J, Shao F. Manipulation of host vesicular trafficking and innate immune defence by Legionella Dot/Icm effectors. Cellular microbiology. 2011;13(12):1870-80.
- Roy CR, Berger KH, Isberg RR. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Molecular microbiology. 1998;28(3):663-74.
- Roy CR, Isberg RR. Topology of Legionella pneumophila DotA: an inner membrane protein required for replication in macrophages. Infection and immunity. 1997;65(2):571-8.
- Marra A, Blander SJ, Horwitz MA, Shuman HA. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proceedings of the National Academy of Sciences. 1992;89(20):9607-11.
- Copenhaver AM, Casson CN, Nguyen HT, Fung TC, Duda MM, Roy CR, et al. Alveolar macrophages and neutrophils are the primary reservoirs for Legionella pneumophila and mediate cytosolic surveillance of type IV secretion. Infection and immunity. 2014;82(10):4325-36.
- Mody CH, Paine III R, Shahrabadi MS, Simon RH, Pearlman E, Eisenstein BI, et al. Legionella pneumophila replicates within rat alveolar epithelial cells. Journal of Infectious Diseases. 1993;167(5):1138-45.
- Waksman G. From conjugation to T4S systems in Gram‐negative bacteria: a mechanistic biology perspective. EMBO reports. 2019;20(2):e47012.
- Li YG, Hu B, Christie PJ. Biological and structural diversity of type IV secretion systems. Protein Secretion in Bacteria. 2019:277-89.
- Liu M, Haenssler E, Uehara T, Losick VP, Park JT, Isberg RR. The Legionella pneumophila EnhC protein interferes with immunostimulatory muramyl peptide production to evade innate immunity. Cell host & microbe. 2012;12(2):166-76.
- Duncan C, Prashar A, So J, Tang P, Low DE, Terebiznik M, et al. Lcl of Legionella pneumophila is an immunogenic GAG binding adhesin that promotes interactions with lung epithelial cells and plays a crucial role in biofilm formation. Infect Immun. 2011;79(6):2168-81.
- Cirillo SLG, Yan L, Littman M, Samrakandi MM, Cirillo JD. Role of the Legionella pneumophila rtxA gene in amoebae. Microbiology (Reading, England). 2002;148(Pt 6):1667-77.
- Newton HJ, Sansom FM, Dao J, Cazalet C, Bruggemann H, Albert-Weissenberger C, et al. Significant role for ladC in initiation of Legionella pneumophila infection. Infect Immun. 2008;76(7):3075-85.
- Rudel T, Boxberger HJ, Meyer TF. Pilus biogenesis and epithelial cell adherence of Neisseria gonorrhoeae pilC double knock-out mutants. Mol Microbiol. 1995;17(6):1057-71.
- Rudel T, Scheuerpflug I, Meyer TF. Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature. 1995;373(6512):357-9.
- Scheuerpflug I, Rudel T, Ryll R, Pandit J, Meyer TF. Roles of PilC and PilE proteins in pilus-mediated adherence of Neisseria gonorrhoeae and Neisseria meningitidis to human erythrocytes and endothelial and epithelial cells. Infect Immun. 1999;67(2):834-43.
- Marko VA, Kilmury SL, MacNeil LT, Burrows LL. Pseudomonas aeruginosa type IV minor pilins and PilY1 regulate virulence by modulating FimS-AlgR activity. PLoS pathogens. 2018;14(5):e1007074.
- Hoppe J, Ünal CM, Thiem S, Grimpe L, Goldmann T, Gaßler N, et al. PilY1 promotes Legionella pneumophila infection of human lung tissue explants and contributes to bacterial adhesion, host cell invasion, and twitching motility. Frontiers in cellular and infection microbiology. 2017;7:63.
- Delves PJ, Martin SJ, Burton DR, Roitt IM. Roitt’s essential immunology: John Wiley & Sons; 2017.
- Sharma AK, Dhasmana N, Dubey N, Kumar N, Gangwal A, Gupta M, et al. Bacterial virulence factors: secreted for survival. Indian journal of microbiology. 2017;57(1):1-10.
- Fischer NL, Naseer N, Shin S, Brodsky IE. Effector-triggered immunity and pathogen sensing in metazoans. Nature microbiology. 2020;5(1):14-26.
- Flor HH. Current status of the gene-for-gene concept. Annual review of phytopathology. 1971;9(1):275-96.
- Patel ZM, Mahapatra R, Jampala SSM. Role of fungal elicitors in plant defense mechanism. Molecular Aspects of Plant Beneficial Microbes in Agriculture: Elsevier; 2020. p. 143-58.
- Colaço HG, Moita LF. Initiation of innate immune responses by surveillance of homeostasis perturbations. The FEBS journal. 2016;283(13):2448-57.
- Dybwad M, Aarskaug T, Fykse EM, Henie Madslien E, Blatny JM. Complete Genome Sequences of Six Legionella pneumophila Isolates from Two Collocated Outbreaks of Legionnaires’ Disease in 2005 and 2008 in Sarpsborg/Fredrikstad, Norway. Genome announcements. 2016;4(6).
- McCloskey A, Perri K, Chen T, Han A, Luo Z-Q. The metaeffector MesI regulates the activity of the Legionella effector SidI through direct protein-protein interactions. Microbes and Infection. 2021:104794.
- Mascarenhas DP, Pereira MS, Manin GZ, Hori JI, Zamboni DS. Interleukin 1 Receptor–Driven Neutrophil Recruitment Accounts to MyD88–Dependent Pulmonary Clearance of Legionella pneumophila Infection In Vivo. The Journal of infectious diseases. 2015;211(2):322-30.
- Barry KC, Ingolia NT, Vance RE. Global analysis of gene expression reveals mRNA superinduction is required for the inducible immune response to a bacterial pathogen. Elife. 2017;6:e22707.
- De Leon JA, Qiu J, Nicolai CJ, Counihan JL, Barry KC, Xu L, et al. Positive and negative regulation of the master metabolic regulator mTORC1 by two families of Legionella pneumophila effectors. Cell reports. 2017;21(8):2031-8.
- Scheiermann J, Klinman DM. Clinical evaluation of CpG oligonucleotides as adjuvants for vaccines targeting infectious diseases and cancer. Vaccine. 2014;32(48):6377-89.
- Oberemok V, Laikova K, Yurchenko K, Marochkin N, Fomochkina I, Kubyshkin A. SARS-CoV-2 will constantly sweep its tracks: a vaccine containing CpG motifs in ‘lasso’for the multi-faced virus. Inflammation Research. 2020;69(9):801-12.
- Lamousé-Smith E, Kelly D, De Cremoux I. Designing bugs as drugs: exploiting the gut microbiome. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2020.
- Rüter C, Schmidt MA. Cell-penetrating bacterial effector proteins: better tools than targets. Trends in biotechnology. 2017;35(2):109-20.
- Jarraud S, Descours G, Ginevra C, Lina G, Etienne J. Identification of Legionella in clinical samples. Legionella: Springer; 2013. p. 27-56.
- Murray PR, Rosenthal KS, Pfaller MA. Medical microbiology E-book: Elsevier Health Sciences; 2020.
- Viasus D, Calatayud L, McBrown MV, Ardanuy C, Carratalà J. Urinary antigen testing in community-acquired pneumonia in adults: an update. Expert review of anti-infective therapy. 2019;17(2):107-15.
- Eble D, Gehrig V, Schubert‐Ullrich P, Köppel R, Füchslin H. Comparison of the culture method with multiplex PCR for the confirmation of Legionella spp. and Legionella pneumophila. Journal of Applied Microbiology. 2021.

This work is licensed under a Creative Commons Attribution 4.0 International License.





