Computational Investigation of Indazole Scaffolds as Tyrosine Kinase Inhibitors Using Molecular Docking and ADMET Prediction
Department of Pharmaceutical Chemistry, School of Pharmacy, Dr. Vishwanath Karad MIT-World Peace University, Pune, Maharashtra, India.
Corresponding Author E-mail: anuruddha.chabukswar@mitwpu.edu.in
DOI : http://dx.doi.org/10.13005/bbra/3013
ABSTRACT:Non-small-cell lung cancer accounted for approximately 85% of newly diagnosed lung cancer cases. In non-small cell lung cancer and tuberculosis, level of vascular endothelial growth factor was found to be elevated; which induces angiogenesis. Many inhibitors are approved for the treatment of non-small cell lung cancer and tuberculosis. However, resistance and severe side effects trigger the search for novel and more potent anti-cancer as well as anti-tuberculosis agents. In this study, molecular docking analysis along with pharmacokinetic ADMET and drug likeness prediction were carried out to evaluate the newly designed indazole scaffolds as potent tyrosine kinase VEGFR-inhibitor. These scaffolds exhibited better binding affinity and favorable interactions with VEGFR-2 enzymes (PDB ID: 4AGD and 4AG8). Out of 10 screened compounds, three most potent compounds (SMO, SBS and SOT) having good scores against 4AGD (-6.99, -6.96 and -6.88 kcal/mol) and compounds (SS, SSA and SMO) having significant scores against 4AG8 (-7.39, -6.71 and -6.70 kcal/mol) emerged as effective and potent VEGFR-2 inhibitors. Based on drug-likeness for oral bioavailability and ADMET risk parameters all the indazole scaffolds may exhibit significant activity. The findings from these current as well as future research efforts will clarify the role of newer indazole derivatives against non-small cell lung cancer and tuberculosis.
KEYWORDS:Dug-likeness; Indazole; Molecular Docking; Vascular Endothelial Growth Factor Receptors
Introduction
Lung cancer is the second most commonly diagnosed cancer worldwide after breast cancer. Every year, it is said to kill a large number of people. The most common type of lung cancer is non-small cell lung cancer (NSCLC), which accounts for 85 percent of all cancer cases1, 2. Angiogenesis is known to be stimulated by abnormal activation of vascular endothelial growth factor receptors (VEGFR), which causes cell proliferation, migration, survival, and permeability of blood vessels3, 4. VEGFRs are divided into three subtypes: VEGFR-1, VEGFR-2, and VEGFR-35. VEGFR-2 inhibitors are small molecules that suppress angiogenesis and lymphangiogenesis by binding to the ATP binding region of VEGFR-26. Furthermore, in vitro and in vivo studies show that VEGF levels are elevated in tuberculosis (TB) patients 7, 8. Aside from the several VEGFR-2 inhibitors that have been approved by the FDA or are in clinical development, there are other initiatives underway to discover novel ones for cancer treatment. Novel drugs having such potential are attempted. Example includes, sunitinib is an indole based antineoplastic agent who inhibits multiple receptor tyrosine kinases9, nintedanib is also an indole based triple angiokinase inhibitor that targets numerous receptor tyrosine kinases and non-receptor tyrosine kinases (nRTKs) 10, 11, urea derivative sorafenib interacts with number of internal (cRAF, BRAF) and cell surface (KIT, FLT-3, VEGFR-2, 3 and PDGFR-β) kinases12, 13, indazole based pazopanib is an second generation multi-targeted tyrosine kinase inhibitor of VEGFR-1, -2 and -3, PDGFR-α, -β and c-KIT14, and another indazole based drug axitinib is an inhibitor of VEGFR-1,-2 and -315, 16.
Indazole scaffolds have been extensively studied for the development of newer pharmaceutical drugs; in particular, numerous indazole derivatives exhibit their capacity to inhibit VEGFR-217, 18. Hence, in this context, our research focuses on the computational design of new indazole based molecules with the pharmacophore characteristics of VEGFR-2 inhibitors. These molecules are made up of various bio-isosters, each of which binds to a distinct area of the ATP binding site19, 20. In this study, molecular docking analysis along with pharmacokinetic ADMET and drug likeness prediction were carried out to evaluate the newly designed indazole scaffolds as potent tyrosine kinase VEGFR-2 inhibitors.
Materials and Methods
This computational work was accomplished on a computer with an Intel Core i5-4570 CPU operating at 3.20 GHz and 4GB of RAM. Software such as Autodock 4.2.6, UCSF Chimera 1.15, ACD/ChemSketch, Biovia discovery studio visualizer, Open Babel GUI 3.1.1, an open chemical toolbox for molecular docking, and pkCSM, a free web tool for ADMET prediction, were used.
Ligand Preparation
Using the ACD/ChemSketch software, a set of ten newly designed compounds with an indazole scaffold was created. All the 3D structures were converted into PDB file format utilizing Open Babel GUI21 and then the energy of all the structures was minimized by assigning 100 steps of steepest descent and 10 steps of conjugate gradient in UCSF-Chimera software 22. All the energy minimized structures were converted into PDBQT file format after detecting root, number of torsions and aromaticity criteria (≤ 7.5) with the help of Autodock tools 23, 24. Two dimensional structures of all the ligands are represented in Table 5.
Receptor Preparation
The 3D crystal structures of VEGFR-2 PDB ID: 4AGD and 4AG8 were retrieved from the protein data bank (PDB) 25. Water molecules, ions, and other ligands present in the protein were removed and the polar hydrogen and Kollman charges were added to the proteins. The PDB format files of proteins were converted into PDBQT file format after assigning AD4 charges. The grid parameter (.gpf) files were prepared by adjusting the size dimension of grid box as 102×100×112 and 102×104×112 (0.5 Å spacing) for 4AGD and 4AG8 proteins respectively. After that the grid log files (.glg) were prepared by launching Autogrid. By utilizing the docking parameter file, input file format (.dpf) docking log file (.dlg) were generated using ADT. The docking investigation was carried out with 10 docking runs for each ligand using the genetic search lamarkain algorithm to explore the optimal conformational space for the ligand. The maximum number of generations and the maximum number of evaluations were both set to 27000 and 2500000, respectively. Finally, the best-fit complexes were analyzed manually using ADT. The discovery studio visualizer was used to visualize and rank the docked receptor and ligand interactions based on binding energy.
Prediction of Drug-likeness and ADMET properties
In this present study, a free online database server, pkCSM (http://biosig.unimelb.edu.au/pkcsm/prediction) was employed to predict the Lipinski’s parameters and ADMET attributes of designed compounds26-28. Topological polar surface area (tPSA) was used to estimate the bioavailability and transportation of an effective drug across the blood-brain barrier 29.
Results and Discussion
Molecular docking studies of designed compounds
All of these newly designed indazole based inhibitors showed negative binding energy (shown in Table 1and 2).
Table 1: Interaction types of newly designed VEGFR-2 inhibitors with 4AGD.
| Entry | Binding affinities
(kcal/mol) |
Conventional hydrogen bonds | Bond distance
(Å) |
Hydrophobic and other interactions |
| SMO | -6.99
|
Glu828, Ile856, Lys826, Arg833 | 1.90, 1.97, 2.40, 2.49 | Leu901, Asp857, Trp827, Phe829, Ala824, Leu902, Trp827, Leu902. |
| SOT | -6.96 | Met806, Ser884, Arg1027 | 1.85, 2.17, 1.90 | Ile888, Leu820, Val805, Leu802, Lys887, Ile888, Lys887. |
| SSPr | -5.50 | Tyr1130 | 2.81 | Thr1131, Pro1128, Thr1132, Tyr938, Ala1127, Tyr1136, Leu1002, Pro937 |
| SPT | -6.09 | Phe918, Ser1037, Arg863, Glu1038, Cys919 | 1.86, 3.39, 2.16, 2.38, 3.39 | Lys1043, Glu917 |
| SMS | -5.96 | Lys1110, Ile1111, Lys1070, Gly1108 | 2.10, 2.20, 2.18, 2.25 | Pro1057, Pro1107, Val1109 |
| SBS | -6.88 | Met806
|
2.28 | Ile890, Leu820, Ile888, Lys887, Leu820, His891, Ile890 |
| SS | -6.54 | Lys826, Leu889, Leu901, Asp857, His894 | 1.81, 2.05, 2.07, 2.24, 2.99 | Ile890, His891, Trp827, Ile890, Leu902, Trp827. |
| SD | -6.32 | Gln1085, Ser1154 | 2.19, 2.90 | Asp807, Ala1020, Thr1152, Ile1084, Lys1023. |
| SSA | -6.17 | Leu902, Lys826, Leu901, Leu889 | 1.84, 2.10, 1.89, 2.21 | Ile890, His894, Trp827. |
| SST | -6.52 | Glu1038, Ser1037, Thr864, Lys838 | 1.98 & 1.99, 2.55, 2.38, 2.57 | Asp852, Glu850, Phe918, Val1041, Lys1043, Cys919, Lys920 |
| Suniti-nib | -8.83 | Leu840, Cys919, Glu917, Lys868 | 1.86, 1.96, 2.08, 2.80 | Asp1046, Val899,Val848 & 916, Phe1047, Phe918, Cys1045, Ala866, Leu1035, Val899 |
Table 2: Interaction types of newly designed inhibitors with 4AG8
| Entry | Binding affinities
(kcal/mol) |
Conventional hydrogen bonds | Bond distance
(Å) |
Hydrophobic and other interactions |
| SMO | -6.70 | Arg842, Leu840 | 1.89, 2.39 | Asp1052, Gly922, Lys1055, Leu840, Leu1035, Cys919, Phe1047. |
| SOT | -6.47 | Arg1027, Ile1025, lys868 | 1.85 & 2.13, 2.22, 2.24 | Cys1024, Asp1026, Gly1048, Cys817, His1026, Ile888. |
| SSPr | -6.45 | Asp998,Tyr1008,
Tyr927, Asn1040 |
2.18, 2.62, 2.44, 3.06 & 2.58 | His1004, Lys997, Ile1036. |
| SPT | -5.82 | Arg1027 | 2.98 | Ala881, Cys817, Cys1024, Val898, Ile1044, Leu1019, Ile888, Ile892, Leu889, Arg1027. |
| SMS | -5.82 | Lys868, Glu885, Asp1046 | 1.84, 2.01, 2.28 | His1026, Gly1048, Leu1019, Val898, Leu889, Ile888. |
| SBS | -6.12 | Lys920, Asn923, Phe921
|
2.06, 2.18 & 2.86, 3.21 | Leu1035, Leu840, Phe1047. |
| SS | -7.39 | Ile1044, Leu1049, Val899 | 2.09, 2.22, 2.26 | Glu885, His1026, Ala881, Cys817, Cys1024, Val898, Ile892, Leu1019, Ile888, Arg1027, Ile1025. |
| SD | -5.92 | Arg1027, Ser884, His1026, Arg819 | 1.86, 1.91, 2.28, 5.56 | Cys817, His1026, Ile888, Cys1024, Ile888, Ile1019, Lys1024. |
| SSA | -6.71 | Ala866, Leu1049, Val914, | 1.88, 2.39, 2.61 & 2.91 | Ile1025, Val899, Leu889, Ile888, Lys868, Val916. |
| SST | -6.61 | Thr1131, Glu1134, Asn1167 | 2.03, 2.48, 2.55 | Asp1129, Tyr938, Pro1133, Ala1127, Ala1166, Ile1006, Leu1163, Tyr1130, Pro937, Leu1002, Thr1132, Glu1134. |
| Axiti-nib | -9.01 | Pro839, Lys919 | 1.91, 1.77 | Lys1045, Val899, Val916, Ala866, Leu1035, Val848, Phe918, Leu840, Phe1047. |
Molecular docking of newly designed VEGFR-2 inhibitors with 4AGD
Based on docking results out of ten compounds screened against 4AGD some of the compounds showed significant binding energies when compared with the native ligand. Compounds showing significant interactions are as follows:
SMO showed the highest binding energy value of -6. 99 kcal/mol and formed four conventional H-bonds with Glu828, Ile856, Lys826 and Arg833 amino acids and bond distances were 1.90, 1.97, 2.40, and 2.49 Å respectively. It formed C-H bond with Leu901 and Asp857 amino acids. It formed π-π staked bond with Trp827 and π-π T-shaped bond with Phe829 amino acid residue. It also formed alkyl bonds with Ala824, Leu902 and π-alkyl bond with Trp827 and a π-lone pair bond with Leu902 amino acid residues of the receptor. Followed by compound SOT showed binding energy value of -6.96 kcal/mol and three conventional hydrogen bond interactions were observed with amino acid positions Met806, Ser884 and Arg1027 and their bond distances were 1.85, 2.17 and 1.90Å respectively. It formed π-σ bonds with Ile888 and Leu820 amino acid residues in addition to H-bonds. It also formed alkyl bonds with Val805, Leu802, Lys887 and π-alkyl bonds with Ile888 and Lys887 amino acid residues. The compound SBS showed the binding energy value of -6.88 kcal/mol formed one conventional H-bond with Met806 amino acid and bond distance was 2.28 Å. It formed π-σ bonds with amino acids Ile890 and Leu820. It also formed amide π-staked bond with Ile890 and alkyl bonds with Ile888, Lys887, Leu820 and a π-alkyl bond with His891 amino acid residues. The binding interaction between designed compounds and the tyrosine kinase VEGFR-2 were shown in Table 1 and Figure 1.
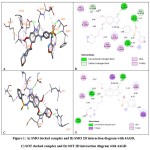 |
Figure 1: A) SMO docked complex and B) SMO 2D interaction diagram with 4AGD, C) SOT docked complex and D) SOT 2D interaction diagram with 4AGD |
Molecular docking of newly designed VEGFR-2 inhibitors with 4AG8
All the newly designed compounds were screened against 4AG8. Compounds having significant interactions are as follows:
Compound SS has the highest binding energy of -7.39 kcal/mol, and forms 3-conventional H-bonds with Ile1044, Leu1049 and Val899 (bond distances of 2.09, 2.22 and 2.26Å respectively) amino acids. It formed carbon hydrogen bonds with the amino acids Glu885 and His1026. It also formed alkyl bonds with Ala881, Cys817 and π-alkyl bonds with Cys1024, Val898, Ile892, Leu1019 and Ile888 residues. It also forms π-cation bond with Arg1027 amino acid residue. It forms one unfavorable bond with Ile1025 amino acid residue. The second best designed compound SSA has binding energy of -6.71 kcal/mol and formed three conventional hydrogen bonds with Ala866 (1.88Å), Leu1049 (2.39Å) and Val914 (2.26 & 2.91Å). It also forms C-H bonds with the Ile1025 amino acid and π-alkyl bonds with Val899, Leu889 and Ile888 amino acid residues. It also forms π-sulfur bond with Lys868 and π-σ bond with Val916 amino acid. The compound SST showed binding energy value of -6.61 kcal/mol and it formed three conventional hydrogen bonds with amino acid Thr1131 (2.03Å), Glu1134 (2.48Å) and Asn (2.55Å). It also formed C-H bond with Asp1129, Tyr938 and Pro1133 and other interactions like alkyl bonds with amino acid Ala1127, Ala1166, Ile1006 and Leu1163. It forms π-alkyl bonds with Tyr1130, Pro937, Leu1002 and Ala1166 amino acid residues. It forms π-σ bond with Thr1132 and one π-anion bond with Glu1134 amino acid residue. Table 2 and Figure 2 indicate the docking scores and amino acids involved in hydrophobic interactions of newly designed compounds against 4AG8. Redocking ligands that were co-crystallized in receptor structures validates the docking calculations (4AGD and 4AG8). In the case of receptor 4AGD, most of the indazole based compounds display significant docking scores as compared to the native ligand sunitinib which has a binding energy -8.83 kcal/mol. When it came to 4AG8, all of the newly designed indazole-based compounds showed a significant binding affinity as compared to the native ligand axitinib, which has a binding energy of -9.01 kcal/mol.
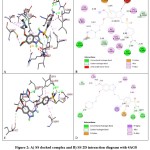 |
Figure 2: A) SS docked complex and B) SS 2D interaction diagram with 4AG8 C) SSA docked complex; D) SSA 2D interaction diagram with 4AG8. |
Prediction of Drug likeness and ADMET properties
All the tested compounds follow Lipinski’s Rule of Five with 0 violations except compound-8 (SD) passes with 2 violations; MW > 500 and rotatable bonds > 10, and compound-9 (SSA) passes with one violation; rotatable bonds > 10 and compound-10 (SST) passes with 3 violations; MW > 500, hydrogen bond acceptor > 10, rotatable bond > 10. The number of H-bond donors and acceptors except compound-10 (SST) was less than 5 and 10, respectively. The property of tPSA was correlated to passive molecular transport across membranes and the blood-brain barrier. Except for compounds SMO, SD, and SST (tPSA greater than 140Å2), all tested compounds pass the GI absorption standard. All the tested compounds were found to have WLOGp values (which predicts whether a molecule has a low toxicity level or not) less than 5. On the scale, these compounds had relatively easy synthetic accessibility (< 5). This means that these compounds are simple to synthesize in the lab and are projected to be active, drug-like, and orally bioavailable. The drug likeness properties of VEGFR-2 inhibitors are represented in Table 3.
Table 3: Drug-likeness properties of newer compounds
| Compd.
Code |
MW | WLOGP | HBD | HBA | RO5 | RB | tPSA | SA |
| SMO | 466.52 | 1.73 | 3 | 9 | 0 | 10 | 152.22 | 3.9 |
| SOT | 460.55 | 2.78 | 2 | 7 | 0 | 9 | 114.16 | 3.61 |
| SSPr | 462.53 | 1.83 | 3 | 8 | 0 | 10 | 139.08 | 3.7 |
| SPT | 460.55 | 2.78 | 2 | 7 | 0 | 9 | 114.16 | 3.6 |
| SMS | 384.54 | 1.04 | 2 | 7 | 0 | 8 | 114.16 | 3.13 |
| SBS | 446.52 | 2.47 | 2 | 7 | 0 | 9 | 114.16 | 3.49 |
| SS | 461.54 | 2.05 | 3 | 8 | 0 | 9 | 140.18 | 3.58 |
| SD | 539.61 | 3.00 | 3 | 10 | 2 | 11 | 151.97 | 3.92 |
| SSA | 489.59 | 1.97 | 3 | 8 | 1 | 11 | 140.18 | 3.55 |
| SST | 579.65 | 3.40 | 3 | 12 | 3 | 13 | 151.13 | 4.4 |
MW: Molecular weight, HBD: Hydrogen bond donar, HBA: Hydrogen bond Acceptor, RO5: Rule of Five, RB: Rotatable bonds, tPSA: Total polar surface area, SA: Synthetic Accessibility.
All of the compounds examined exhibit good GI absorption values (all above 66%) and passed the 30% criterion, indicating that these compounds have high human intestine absorption capabilities. All the tested compounds served as Pgp substrates. These tested VEGFR-2 inhibitors were found to have low skin permeability (logkp > -2.5) and compounds showed moderate water solubility. The BBB permeability (logBB) values of all compounds were < 0.3, which indicates that these inhibitors are poorly absorbed across the brain. The CNS permeability (Log PS) values for all were > -3, which implies that all these compounds can penetrate the CNS. Moreover, they were found to be substrates for all the CYP isoforms except CYP 3A4. The compounds SOT, SPT, SBS, SD, SSA and SST were the substrates and compounds SOT, SPT, SBS, SD and SST were found to be inhibitors of CYP 3A4. Furthermore, in the body, a drug molecule’s total clearance was within permissible limits. All these newly designed VEGFR-2 inhibitors were found to be non-toxic, except for compound SS. According to these results, the newly designed VRGFR-2 inhibitors have a high absorption value, low toxicity level, and good cell membrane permeability. All of these newer VEGFR-2 inhibitors were predicted to have favorable pharmacokinetic properties. Table 4 shows the predicted ADMET properties of all of these compounds.
Table 4: ADMET properties of designed compounds
|
Compd. Code |
ABS | Distribution | Metabolism |
Excretion
TC |
AMES Toxicity |
|||||||
|
Int. Abs. (%) |
LogBB
|
LogPS |
CYP Substrate | CYP Inhibitors | ||||||||
| 2D6 | 3A4 | 1A2 | 2C
19 |
2C9 | 2D6 | 3A4 | ||||||
| SMO | 66.697 | -2.160 | -4.591 | N | N | N | N | N | N | N | 0.63 | N |
| SOT | 77.107 | -1.616 | -3.752 | N | Y | N | N | N | N | Y | 0.79 | N |
| SSPr | 67.186 | -2.066 | -4.555 | N | N | N | N | N | N | N | 0.74 | N |
| SPT | 80.077 | -1.631 | -3.760 | N | Y | N | N | N | N | Y | 0.89 | N |
| SMS | 74.651 | -1.611 | -4.385 | N | N | N | N | N | N | N | 0.92 | N |
| SBS | 79.609 | -1.604 | -3.801 | N | Y | N | N | N | N | Y | 0.88 | N |
| SS | 72.529 | -1.682 | -3.937 | N | N | N | N | N | N | N | 1.01 | Y |
| SD | 79.068 | -2.217 | -4.239 | N | Y | N | N | N | N | Y | 0.84 | N |
| SSA | 66.197 | -1.796 | -3.800 | N | Y | N | N | N | N | N | 1.20 | N |
| SST | 72.848 | -2.334 | -4.263 | N | Y | N | N | N | N | Y | 1.17 | N |
ABS: Absorption, Int. Abs: Intestinal Absorption, TC: Total Clearance
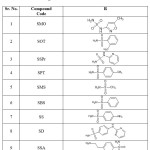 |
Table 5: 2D structures of the newly designed VEGFR-2 inhibitors. |
Conclusion
This work addresses, the molecular docking simulation carried out on indazole based newly designed compounds as VEGFR-2 inhibitors. In this study, compounds SMO (-6.99 kcal/mol) docked with 4AGD and compound SS (-7.39 kcal/mol) docked with 4AG8 had the highest binding affinity. A significant binding affinity score was found in the majority of newly designed compounds. Drug-likeness and ADMET prediction concluded that these compounds are orally bioavailable with high absorption, less toxic and high permeable properties, and most of the compounds follow Lipinski’s Rule of Five. Furthermore, these indazole scaffolds were discovered to have good synthetic accessibility (<5) indicating that they are easy to synthesize in the lab. The results observed in the present study demonstrated that, after further refinement, the newly designed indazole based compounds could be the potential drug of choice for the treatment of NSCLC and tuberculosis.
Acknowledgement
All authors are thankful to the management of Dr. Vishwanth Karad, MIT-World Peace University, Pune, India for the support. (Formerly MAEER’s Maharashtra Institute of Pharmacy, Pune-411038).
Conflict of Interest
The authors declared no conflict of interest.
Funding Source
Authors are grateful to DST-SERB for financial support of this research.
References
- Ching Yao Yang, James Chih H, sin Yang and Pan-Chyr Yang. Precision management of advanced Non–Small Cell Lung Cancer. Rev. Med., 2020; 71:117–36.
CrossRef - Kong L L, Ma R, Yao M Y, Yan X E, Zhu S J, Zhao P, Yun C H. Structural pharmacological studies on EGFR T790M/C797S. Biochem. Biophys. Res. Commun., 2017; 488(2):266–272.
CrossRef - Hicklin DJ and Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. Clin. Oncol., 2005; 23(5):1011–1027.
CrossRef - Richard D. Hall, Tri M. Le, Daniel E. Haggstrom, Ryan D. Gentzler. Angiogenesis inhibition as a therapeutic strategy in non-small cell lung cancer (NSCLC). Transl. Lung Cancer Res., 2015; 4(5):515-523.
- Olsson A.K., Dimberg A, Kreuger J, Claesson Welsh L. VEGF receptor signaling-in control of vascular function. Rev. Mol. Cell Biol., 2006; 7:359–71.
CrossRef - Olmes K, Roberts O. L, Thomas A. M, Cross M. J. Vascular endothelial growth factor receptor-2: structure, function, intracellular signalling and therapeutic inhibition. Cell Signal, 2007; 19:2003–12.
CrossRef - Polena H. et al. Mycobacterium tuberculosis exploits the formation of new blood vessels for its dissemination. Rep., 2016; 6:33162.
CrossRef - A Spagnuolo, G Palazzolo, C Sementa & C Gridell. Vascular endothelial growth factor receptor tyrosine kinase inhibitors for the treatment of advanced non-small cell lung cancer. Expert Opin. Pharmacother., 2020; 21(4):491-506.
CrossRef - Alfredo Carrato Mena, Enrique Grande Pulido and Carmen Guillen-Ponce.Understanding the molecular-based mechanism of action of the tyrosine kinase inhibitor: sunitinib. Anti-Cancer Drugs, 2010; 21:S3–S11. .
CrossRef - Roth GJ, Heckel A, Colbatzky F. Design, synthesis, and evaluation of indolinones as triple angiokinase inhibitors and the discovery of a highly specific 6-methoxycarbonyl-substituted indolinone (BIBF 1120). Med. Chem., 2009; 52(14):4466–80.
CrossRef - Hilberg F, Roth GJ, Krssak M. BIBF 1120: Triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy. Cancer Res., 2008; 68(12):4774–82.
CrossRef - Wilhelm SM, Adnane L, Newell P, Villanueva A, L lovet JM, Lynch M. Preclinical overview of sorafenib, a multikinase inhibitor that targets both Raf and VEGF and PDGF receptor tyrosine kinase signaling. Cancer Ther., 2008; 7(10): 3129–40.
CrossRef - Keating GM, Santoro A. Sorafenib: a review of its use in advanced hepatocellular carcinoma.Drugs., 2009; 69(2): 223–40.
CrossRef - Sonpavde G, Hutson TE, Sternberg CN. Pazopanib, a potent orally administered small-molecule multitargeted tyrosine kinase inhibitor for renal cell carcinoma. Expert Opin. Investig. Drugs., 2008; 17(2):253-61.
CrossRef - Cohen EE, Rosen LS, Vokes EE, Kies MS, Forastiere AA, Worden FP, Kane MA, Sherman E, Kim S, Bycott P, Tortorici M, Shalinsky DR, Liau KF, Cohen RB. Axitinib is an active treatment for all histological subtypes of advanced thyroid cancer: results from a phase II study. Clin. Oncol., 2008; 26(29):4708-13.
CrossRef - Gross-Goupil M, Francois L, Quivy A, Ravaud A. Axitinib: a review of its safety and efficacy in the treatment of adults with advanced renal cell carcinoma. Med. Insights Oncol., 2013;7:269-77.
CrossRef - Dvorak HF. Vascular permeability factor/vascular endothelial growth factor: a critical cytokine in tumor angiogenesis and a potential target for diagnosis and therapy. Clinl. Oncol., 2002; 20:4368-80.
CrossRef - Joukov V, Kumar V, Sorsa T. A recombinant mutant vascular endothelial growth factor-C that has lost vascular endothelial growth factor receptor-2 binding, activation, and vascular permeability activities. Biol. Chem., 1998; 20:6599-602.
CrossRef - Abdel Mohsen HT, Abdullaziz MA, Kerdawy AME. Targeting receptor tyrosine kinase VEGFR-2 in hepatocellular cancer: rational design, synthesis and biological evaluation of 1, 2-disubstituted benzimidazoles. Molecules, 2020; 25:770.
CrossRef - Rampogu S, Baek A, Park C. Discovery of small molecules that target vascular endothelial growth factor receptor-2 signalling pathway employing molecular modelling studies. Cells, 2019; 8:269.
CrossRef - N O’Boyle. M Bank, C A James, C Morley, T Vandermeersch and G R Hutchison. Open Bable: An open chemical toolbox. Cheminform., 2011; 3:33.
CrossRef - Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-visualization system for exploratory research and analysis. Comput. Chem., 2004; 25(13):1605-12.
CrossRef - Forli, S., Huey, R., Pique, M. E., Sanner, M. F., Goodsell, D. S., & Olson, A. J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Protoc., 2016; 11(5): 905–919.
CrossRef - Ferreira, L., dos Santos, R., Oliva, G., & Andricopulo, A. Molecular Docking and Structure-Based Drug Design Strategies. Molecules, 2015; 20(7):13384–13421.
CrossRef - McTigue M., Murray B. W., Chen J. H., Deng, Y. L., Solowiej, J.& Kania R. S. Molecular conformations, interactions, and properties associated with drug efficiency and clinical performance among VEGFR TK inhibitors. Natl. Acad. Sci. U.S.A., 2012; 109(45):18281–18289.
CrossRef - Lipinski C. A., Lombardo F., Dominy B. W. & Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Drug Deliv. Rev., 2001; 46(1-3):3–26.
CrossRef - Daina A., Michielin O. & Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Rep., 2017; 7(1):42717.
CrossRef - Ertl, P., Schuffenhauer A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. Cheminform., 2009; 1:8.
CrossRef - Ertl, P., Rohde, B. & Selzer, P. Fast. Calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. Med. Chem., 2000; 43:3714–3717.
CrossRef
Accepted on: 23-07-2022
Second Review by: Dr. Shahina Akter
Final Approval by: Dr. Ghulam Md Ashraf









