How to Cite | Publication History | PlumX Article Matrix
Potential of Molecular Plant Breeding for Sustaining the Global Food Security
Ritu Mahajan* and Nisha Kapoor
and Nisha Kapoor
School of Biotechnology, University of Jammu, Jammu and Kashmir, India
Corresponding Author E-mail: ritufeb@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3072
ABSTRACT: New alleles are continuously added to bring novel traits which are selected using genetic manipulations. Understanding the genes responsible for a particular phenotype involves recent genomic approaches which are to be integrated with conventional breeding programs for the crop improvement. Use of biotechnological tools merged with the conventional breeding practices has resulted molecular plant breeding which has significant contributions to food production, despite the presence of narrow genetic base in current materials used for breeding purposes. Use of molecular markers along with high-throughput genome sequencing efforts, have dramatically augmented our information to characterize the elite germplasm for the production of hybrids and improved populations. This review focuses on possibilities for the need and application of molecular breeding tools in the genetic improvement of the crop plants which can ensure sustainable food production for the increasing global population.
KEYWORDS: Genetic Diversity; Marker Assisted Selection; Molecular Markers; Plant Breeding, Sequencing
Download this article as:| Copy the following to cite this article: Mahajan R, Kapoor N. Potential of Molecular Plant Breeding for Sustaining the Global Food Security. Biosci Biotech Res Asia 2023;20(1). |
| Copy the following to cite this URL: Mahajan R, Kapoor N. Potential of Molecular Plant Breeding for Sustaining the Global Food Security. Biosci Biotech Res Asia 2023;20(1). Available from: https://bit.ly/3HQpXgC |
Introduction
Agriculture being dominant in the world today, hunger and malnutrition continue even in developing countries. With the increase in the global population, the rate of food production needs to be doubled, so as to feed ten billion people by the year 2050 1. Although, the growth rate of the crop yield is 2.4% per annum but still the yield of major food crops or cash crops is low 2, and still the increase in crop production recorded is far less than half the required rate. Over a century, a remarkable progress has been made in plant breeding with the mandate to increase the crop yield and this was made possible due to the huge developments in the fields of agricultural technology. Since, most of the traits in breeding programs are quantitative in nature so the selection in classical breeding is based on phenotype rather than genotype and this result in the selection of favorable genes in the course of time 3. Plant breeding has the ability to alter the traits of plants where the desired characteristics related to specific traits are selected and this results in the production of certain elite varieties with superior agricultural characters.
To feed the world population, enhancement by 70% in the current food production is required, but mostly due to unfavorable consequences of climatic changes and variations, the crop yield is badly affected thus causing a threat to food security 4. Also, with time, the agricultural land and water resources are becoming stagnant and thus that the desired increase in crop production can be attained by eco-efficient crop production systems. However, the major challenge still lies in doubling the food production but at the same time without compromising with environmental integrity and public health. Thus, plant breeding needs re-orientation so as to generate ‘smart’ crop varieties that have higher yield with fewer inputs. Such crop plants can only be raised by the application of advanced molecular tools 5,6. This will involve the re-engineering of plants with desirable traits and genetic stability along with the increase in food productivity by two to three folds.
In this review, we have discussed few molecular based methods that could be integrated into traditional plant breeding practices (Fig 1) to meet the food productivity challenges. Molecular based tools and marker-assisted selection has resulted in decrease in the time required for developing a new crop variety with desirable traits. Several earlier reviews have demonstrated the use of molecular markers in crop improvement but their reliability/reproducibility as selection criteria for various plant populations was still not clear. The present review focuses on the recent advancements made in the field of molecular biology and how they are used for enhancing the crop productivity either by introducing the new genes/alleles or by editing the genome therby opening new opportunities for basic research in the fields of plant biology (Table 2).
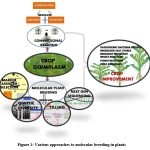 |
Figure 1: Various approaches to molecular breeding in plants. |
Forward and reverse mutagenesis
Plant variations occur due to mutations and crossings and in their absence, conventional breeding based on phenotypic selection would be not possible in the nature. Mutations are the primary source of genetic variations that provides the raw material for selection and is also a powerful force playing significant role in evolution 7. Spontaneous mutations are uncommon with low mutation rate and since they occur naturally so it’s difficult to utilize them in the plant breeding programs. Hence, certain constraints that occur in conventional breeding like low vigour and fertility, complex genomes, narrow gene pool, lengthy breeding cycle can be addressed through induced mutagenesis. Apart from them, transitions and transversions induce variations in base pair changes while additions and deletions results in alteration in reading frame while duplications change the gene sequence and its functioning. Thus, development of new varieties through mutation breeding along with recombination has added new traits to the elite germplasm.
The main aim for mutation-based breeding is to increase the number of new varieties or to improve already adapted plant varieties, which is generally done by modifying one or two major traits to increase the productivity or quality of plants 8. Plant mutagenesis has introduced variations in barley crop plants where ethylmethanesulfonate (EMS) based mutated plant populations were screened for two genes followed by PCR-based mutation scanning and detection of dye labeled cleaved products 9.
Mutation-based breeding suggests a unique opportunity to recognize the novel traits in the population but at the same time, for complex traits, these variations could not be screened using simple genotypic or phenotypic screening methods. Thus, muta-genomics is becoming an essential tool where mutant traits are investigated using high throughput technologies for analyzing gene functions 10. Several mutagenesis methods like transposon tagging along with T-DNA insertion and ionizing radiation have offered an important knowledge about the type of mutations 11. Insertional mutagenesis or transposons results mainly in disordering the gene sequence as compared to EMS based chemical mutagenesis that results in point mutations 12.
Introducing chemical based mutations in the development of TILLING (Targeting Induced Local Lesions in Genomes) technology has resulted in the introduction of random mutations across the entire genome. TILLING involves extraction of genomic DNA from each mutated line and screening of the plant population using advanced molecular techniques 13. TILLING technique has been applied successfully to barley, rice, wheat, maize, oat, pea and soybean 14,15. Chawade et al.16 developed a TILLING population of spring oat, which has a mutation frequency of one mutation per 20-40 kb and several mutations in some important genes that could develop oat resources with specific traits. Plant mutant repositories are being established as in barley HorTILLUS (Hordeum—TILLING—University of Silesia) platform, which is the largest and an eternal source for new mutations for barley researchers where a mutagenized population was created for spring barley cultivar with majority of transitions and few for transversions 17. Thus, this technology is a combination of both high frequency of induced mutations and receptive techniques for discovering point mutations in the plant populations 18.
Germplasm variation and genetic diversity
The foundation for molecular plant breeding was shaped by the use of genetic manipulations at DNA level so as to improve plants with desired characters. This involves the integration of advanced biotechnological techniques along with the molecular marker applications. Without the introduction of DNA markers, the idea of discovering the loci controlling complex traits was impossible. Currently, using molecular markers these genome regions can be easily flagged, thereby allowing the selection for Quantitative trait loci (QTLs) (Table 1).
Increase in population pressure and expansion in agricultural land has resulted in the erosion of plant biodiversity which ultimately lead to the large-scale extinction of agriculturally important crop species. After pandemics such as the famous Irish potato famine and Southern corn leaf blight epidemic in USA, crop varieties with higher yields were introduced under green revolution to overcome the food crisis. As a result, many gene banks were initiated by the Consultative Group of International Agricultural Researches (CIGAR) and few centers for exploring research in conserving the genetic resources for various food crops around the world.
Genetic diversity is the foundation to biodiversity but inbreeding among plants has resulted in recombination between undesirable genes 19, 20. Use of molecular tools in plant breeding resulted in expansion of genetic diversity and also provides opportunity to farmers and breeders in developing new varieties with improved characteristics. Though, the phenotypic variation has positive impact on genetic diversity, yet it is dependent upon the interactions between genotype and environmental factors 21. This is due to the presence of variations in the phenotype which are maintained further in the successive generations and sometimes results in the origin of new plant species thus contributing to the genetic diversity 22.
Since, the genetic diversity changes over time under artificial selection hence more efforts are required to explore the crop genetic diversity so as to attain sustainable crop production in near future 23. Domestication, natural selection and mutations have great impact on genetic diversity. Domestication reduces, while mutations increase the genetic diversity. Similarly, directional selection decreases while at the same time disruptive selection increases the genetic diversity 24.
Recent molecular techniques such as New Generation Sequencing, has gained importance due to its low cost, less time for sequencing the genome and the accumulation of huge data. Several software programs are accessible for estimating diversity at phenotypic and molecular level thus increasing the effectiveness of germplasm for crop improvement.
MAS for quantitative trait selection
Most of the traits in breeding programs are quantitative in nature; while in classical breeding selections are made on the basis of phenotype without taking into account the genes that gets selected in each consecutive generation 3. Hence, enhancement in the crop yield under conditions such as water scarcity, degradation of arable land, pollution, biotic and abiotic stresses along with consumer preferences, has become a challenge for plant breeders and scientists to develop new crop varieties persistently.
With the arrival of molecular markers the likelihood of selecting the desirable traits has became easier and are used frequently in the plant breeding programs.. Marker-assisted selection (MAS), where the molecular markers are closely linked to gene of interest, has now a days become a feasible tool to select a particular trait in the breeding programs for several crop species 25,26,27. Using associated markers, it is interpreted that MAS depends upon the genotype of plants rather than its phenotype, so it is an indirect selection of crops with improved traits based on the closely associated markers 28, 29, and once the association between marker and trait is linked then the desirable traits can be transferred from parents to offsprings through marker assisted backcrossing 30.
The main challenge faced by breeders is where introgression and multiple genes pyramiding (for polygenic traits) affecting the same trait is to be done in short time. Since gene pyramiding involves combinations of various desirable genes so it is impossible through conventional breeding. However, adapting a resistant crop using gene pyramiding is the most desirable strategy to manage diseases 31. Hence, MAS bridges the gap in crop development and also overcome big challenges and predictions of conventional breeding. In Africa, MAS has resulted in the recognition of quantitative trait loci (QTLs) that affects the productivity and nutritional quality of maize. Also many potential QTLs linked to good agronomic traits have been mapped which show better performance in MAS breeding programs 32.
Diversification of gene pool using exotic germplasm
Widening the gene pool using exotic sources, can be challenging, as it leads to the introduction of many unwanted traits/alleles. Breeders have successfully used exotic germplasm for harnessing the germplasm by introducing the potential alleles associated with several desirable traits. The choice of the donor parents is prime factor to introgress alleles for specific attributes that can increase the crop productivity otherwise linkage disequilibrium can have deleterious effect 33. Similarly, use of allied species can result in sterility of hybrids and linkage drag during interspecific hybrid production, 34. In Brassica the elite breeding lines are recommended for canola cultivar development that has high-oleic acid content 35. This makes the identification of right allelic combinations a difficult task. Sequencing of Brassica genome has facilitated the discovery of favorable alleles and their introduction into existing breeding material using biotechnological tools 36. Similarly, on analyzing the diversity of B. napus gene pools with identified but allied Brassica species, genetic differences in B. napus were observed and thus these new favorable alleles can be introduced by the breeders to create better allelic combinations in Brassica. Oilseeds Research Institute, Faisalabad developed a new variety called “Super canola” which is high yielding and disease resistant. This variety was developed from RBN-03052 (a local canola line of rapeseed which is high yielding) and an Australian rapeseed variety (Rainbow), that has low Erucic acid and Glucosinolates 37.
Introgression of genes from wild/distant relatives
Mostly in some crop plants, the wild relatives serve as donors of useful genes for the genetic improvement of their progeny 38,39. For this the genetic value of wild relatives needs to be conserved and can be further enhanced by the implementation of new breeding techniques. Even the information gained from model plant species also promises in the conservation of these genetic resources and transfer of the favorable alleles to generate new crop varieties. However, the environment from time to time poses problems for the recognition of positive alleles by hampering the expression of these useful alleles. Even the transfer of alleles into breeding material for generating the new crop varieties is not cost effective process as several plants with undesirable characteristics need to be eliminated at early stages of development so that only a few crop lines remain for testing 40. In sunflower, genes for cytoplasmic male sterility, herbicide tolerance and biotic stress resistance have been successfully introgressed from the wild relatives into the cultivated gene pool, without any decrease in oil content and its quality 41. Similarly, in rice, genes for bacterial blight resistance have been transferred from wild rice O. nivara and O. glaberrima to its cultivated variety O. sativa 42,43 (Table 1). This transfer of new alleles/gene(s) from wild resources to cultivated varieties will help in broadening the genetic base of the crop plants in near future.
Biofortification for nutrition improvement
Molecular breeding being a cost-effective tool contributes to global food security by increasing yield and the nutritional value of forage and crop plants. Biofortification, which is a new strategy for the improvement of crop nutritional quality through breeding addresses micronutrient and vitamin B6 deficiencies worldwide especially in women and children below 5 years 44,45. It involves the development of micronutrient-rich staple crops using traditional breeding and modern biotechnology tools. Recent surveys in India have unraveled the positive effects of biofortified maize and wheat on young children. The consumption of zinc-biofortified wheat in form of chappatis, flatbread and porridge by rural adults and children has increased their immunization against pneumonia and vomiting generally caused by conventional wheat products 46.
The Consultative Group on International Agricultural Research (CGIAR) is exploring the potential to enhance the bioavailablity of iron and zinc content in staple crops such as rice, wheat and maize 47,48. Beasley et al. 49 enhanced iron and zinc content in the wheat grains by transferring rice nicotianamine synthase-2 gene to upregulate the biosynthesis of nicotianamine and 2-deoxymugineic acid. Quality protein maize, a biofortified maize was introduced by the International Maize and Wheat Improvement Center, CIMMYT due to mutation in the opaque-2 gene that resulted in enhanced levels of essential amino acids (lysine and tryptophan) which were earlier lacking in the proteins of maize endosperm and hence reduced malnutrition in young children 50. Similarly, millets are an important source of energy next to cereals, due to the presence of high protein content, essential amino acids, minerals and vitamins in their grains. HarvestPlus (a Program of the Consultative Group on International Agricultural Research), has released millet with high iron content in India to overcome the iron deficiency. The group decoded the QTLs and the genes linked to protein quality in finger millet using comparative genomics 51. Thus, enhancing the nutrient value of crops through biofortification is a cost effective approach and further needs institutional leadership along with public and private policies to achieve the goals.
Genetic transformation
Favorable nutrition plays an important role in human health and progression. Crop yield is severely affected due to adverse environmental conditions. Thus, a major goal is to find different ways to increase the productivity as well as nutritional value in crop plants. The main crisis in 21st century agriculture is to attain 70% enhanced crop productivity in coming years in spite of the foreseen unfavorable environmental conditions so as to sustainably feed human population 52,53.
Genetic engineering is a ray of hope for improving agricultural productivity. Using this technique, improvement in crop can be done for enhanced nutritional value, tolerance to stresses, and efficient use of soil nutrients and water. Several transgenic crops such as rice, wheat, maize, cotton, potato, canola and sugar cane are growing globally and countries like US and Brazil are their leading producers.
Transgenic crops with novel candidate genes such as stress-responsive NAC transcription factors in rice, Alfin-like gene from Atriplex hortensis in Arabidopsis against drought and salt stress provide a quick method to develop stress tolerant crop varieties 54,55. Similarly, in wheat transgenic, NF-YA transcription factor related to low nitrogen and low-phosphorus-inducible was over-expressed which resulted in significant increase in nitrogen and phosphorus uptake 56 along with a second transcription factor that played a role in nitrogen signaling 57, thus resulting in improved grain yield.
Similarly, there are several approaches that can increase the gene expression levels by the random activation of endogenous genes using enhancers, have conferred new functions to the plants 58. Studies have revealed that a random insertion of transcriptional enhancers into the genome using strong promoter can result in over-expression of a member of a gene family. This involves no interference from other gene families, thus resulting in a novel phenotype and characterization of functionally redundant genes 59.
Next-generation sequencing and gene identification
Recently, advances in high-throughput genome sequencing have dramatically augmented the capability to characterize the genetic variants present in the germplasm including minor crops 60. In finger millet, high-throughput sequencing has resulted in the characterization of calcium sensing and its accumulation mechanisms across genotypes and also to untangle the physiological and molecular basis of salinity response among genotypes under greenhouse conditions 61,62. There information enabled the identification and maintenance of beneficial alleles and better choices of parents for producing hybrids.
Next generation sequencing has transformed the field of plant breeding by developing highly polymorphic, cost effective and closely linked molecular markers for a target trait 63. Also, it led to the development of genome wide sequence based markers, which are less costly, both for minor and non-model crop plants. This has also facilitated the development of genomic resources for characterization of diversity, gene discovery, evolutionary relationships and marker assisted breeding in orphan crops which have no prior information on its genomic sequence 64.
Genome Editing
Genome-editing tools along with nucleases that are site-specific have provided efficient targeted modification in plant systems 65. Genome-editing techniques have been utilized globally as an effective plant breeding tool against large variety of crop species 66. During early times, techniques like use of zinc-finger nucleases 67 and transcription activator-like effector nucleases 68 were used for editing, but recently development of clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system) has re-organized the editing efficiency with a variety of potential applications that ranges from gene knockouts to the introduction of small indels causing frame-shift mutations or introduction of premature stop codons 69.
CRISPR technique apart from being simple, efficient, and cost effective has also ability to target multiple genes. The CRISPR cleavage requires a short synthetic guide RNA sequence (about 20 nucleotides) that binds to the target DNA and a Cas9 nuclease enzyme that cleaves 3–4 bases after the protospacer adjacent motif (PAM). The Cas9 nuclease is made up of two domains, RuvC-like domains and a HNH domain, and each domain cut one DNA strand 70. CRISPR/Cas9 techniques have been used widely against several plant genomes and the majority of editing has been done in plant species such as Arabidopsis, rice and tobacco 71.
In rice, reports have revealed that CRISPR/Cas9 targeted mutation occurring in the ethylene responsive factor increased resistance to blast disease 72, while development of two knockout mutants of OsSWEET13 enhanced its resistance to bacterial blight disease 73. Multiplexed genome editing was also done in hexaploid wheat where genes, one for grain trait and two for disease resistance were targeted using three gRNAs and a single promoter and further the editing efficiency was tested in transgenics 74. In maize, Zhu et al. 75 using maize promoter (U6 snRNA), edited phytoene synthase gene involved in carotenoid biosynthesis that resulted in white kernels and albino seedlings. Transgenic showed that only stable albino mutants were produced without any off-target sites edited. Similarly, Young et al. 76 used a three-step approach in maize where computational prediction was combined with biochemical detection. They studied that if promiscuous guide RNA is intentionally designed with mismatches and in case if one mismatch occurrs in the PAM proximal region then only predictable off-target edits could be observed and minimized.
Thus, CRISPR/Cas9 based genome editing technique quickly inserts useful traits and improves agronomic output. This technique can help in designing suitable crops that will adapt to the current environmental conditions and will feed the growing human population to overcome global hunger.
Table 1: Molecular breeding for various traits in crop plants.
|
S. No |
Crops |
Traits |
References |
|
1
|
Rice
|
Blast disease resistance |
Li et al. 2019, Ning et al. 2020 |
|
Genetic diversity |
Hour et al. 2020; Hassan et al. 2022 |
||
|
Nicotinamide synthase gene and Ferritin Gene |
Boonyaves et al. 2017 |
||
|
Yield enhancement |
Gaikwad et al. 2014 |
||
|
Bacterial blight resistance |
Pradhan et al. 2015; Kumari et al. 2020 |
||
|
2
|
Wheat
|
Rust resistance genes |
Yang et al. 2017; Babu et al. 2020 |
|
Iron and zinc concentration |
Wani et al. 2022 |
||
|
Osmotic stress tolerance gene |
Yang et al. 2016 |
||
|
3 |
Maize
|
Downy mildew resistance genes |
Lohithaswa et al. 2015 |
|
Provitamin A |
Zunjare et al. 2018 |
||
|
4
|
Sunflower
|
Drought resistant genes |
Hussain et al. 2017 |
|
Heat and Drought stress genes |
Killi et al. 2017 |
||
|
5 |
Finger millet |
Salinity gene |
Singh et al. 2022 |
|
6 |
Soybean |
Seed oil content |
Yao et al. 2020 |
Table 2: Advantages and disadvantages of various techniques used in molecular breeding.
|
Techniques |
Advantages |
Disadvantages |
|
Forward and reverse mutagenesis
|
Alternation in protein by the mutant allele |
Sometimes deleterious modifications as it is not a targeted approach |
|
Genetic diversity |
Strengthens the ability of populations to resist biotic and abiotic stresses |
Accumulation of number of deleterious genetic variations, which may increase the risk of extinction of a population |
|
Marker Assisted Selection |
Increases the efficiency for breeding as compared to conventional breeding |
MAS is more expensive than conventional techniques |
|
Gene introgression from wild relatives |
Wild plants are source of desirable traits for disease resistance, fruit quality, and abiotic stresses
|
Linkage drag resulting in transfer of undesirable genes |
|
Biofortification |
Increase in food productivity and quality |
Over-consumption of the nutrients |
|
Genetic transformation |
Increased food supply with less cost and longer shelf life |
Addition of new allergens in food |
|
Next Generation Sequencing |
Generation of genomic resources in plants and decoding of a species genome |
To differentiate various paralogous genes and pseudogenes |
|
Genome editing |
Crops are designed to make them more resistant to pests, reduction in pesticide use |
Chance of errors is more during the gene editing process |
Conclusions and Future Prospects
Promising food security along with providing healthy food is a key challenge of the present day. Plant breeding has made an outstanding development in crop improvement by combining molecular techniques with conventional breeding that has created a wealth of information which can be further exploited for genetic improvement of crop plants, though an understanding of the current barriers and appropriate solutions needs to be developed. Though, breeding allows the pre-selection of genotypes with the desired combinations of alleles before field testing but still genetic constraints like epistasis, linkage drag of unwanted alleles remains challenge for cultivar development. The final aspire of molecular plant breeding is to introduce desirable traits in crop plants through certain breeding methods like backcrossing and recurrent selection, thus reducing the time for conventional selection which is based exclusively on phenotype. Further, advanced biotechnological tools and the genomic resources are providing plant breeders with new techniques and methodologies. Also, the beginning of high-throughput sequencing and genotyping, along with molecular tools has proved to be a boon for developing and under developed countries for dissecting the complex traits that exists due to strong environmental influence in the staple crops. However, there is still need to develop strategies using modern genomics tools and breeding approaches in combination for the successful integration of desirable genes in crops against destructive pathogens, biotic and abiotic stresses for the production of food in the near future.
Acknowledgement
The authors thankfully acknowledge various funding agencies such as JK-DST, DST, DBT-GoI, UGC, NMPB for financial aid in the form of major research projects and School of Biotechnology, University of Jammu, Jammu. In addition, the authors are thankful to RUSA, SAP and PURSE grants, Central Facility and Bioinformatics Center at School of Biotechnology, University of Jammu for their support.
Conflict of Interest
The authors declare that they have no conflict of interest.
Funding Sources
There are no funding source.
References
- Fróna D, Szenderák J, Harangi-Rákos M. The Challenge of Feeding the World. Sustainability. 2019; 11:5816.
CrossRef - Eliazer Nelson ARL, Ravichandran K, Antony U. The impact of the green revolution on indigenous crops of India. Ethn. Food. 2019; 6:8.
CrossRef - Bassi FM, Bentley AR, Charmet G, Ortiz R, Crossa Breeding schemes for the implementation of genomic selection in wheat (Triticum spp.). Plant Sci. 2016;242:23-36.
CrossRef - Shahzad A, Ullah S, Dar AA Sardar MF, Mehmood T, Tufail MA, Shakoor A, Haris M. Nexus on climate change: agriculture and possible solution to cope future climate change stresses. Environ Sci Pollut Res Int. 2021; 28:14211-14232.
CrossRef - Sarfraz Z, Iqbal MS, Pan Z. et al. Integration of conventional and advanced molecular tools to track footprints of heterosis in cotton. BMC Genomics. 2018;19:776.
CrossRef - Mahajan R, Kapoor N. Molecular Breeding Strategies for Genetic Improvement in Rice (Oryza sativa). In: Advances in Plant Breeding Strategies: Cereals, J. M. Al-Khayri et al. (eds.), Springer Nature Switzerland AG. 2019 pp. 317-342.
CrossRef - Holme IB, Gregersen PL, Brinch-Pedersen Induced genetic variation in crop plants by random or targeted mutagenesis: convergence and differences. Front. Plant Sci.2019; https://doi.org/10.3389/fpls.2019.01468.
CrossRef - Srivastava PS, Pandey M, Tiwari DK. Mutagenic effects of sodium azide on the growth and yield characteristics in wheat (Triticum aestivum em. Thell.). Asian J. Plant Sci. 2011;10:190-201.
CrossRef - Caldwells DG, McCallum N, Shaw P, Muehlbauer GJ, Marshall DF, Waugh RA. Structured mutant population for forward and reverse genetics in barley (Hordeum vulgare) Plant J. 2004; 40:143–150.
CrossRef - Nawaz Z, Shu Q. Molecular nature of chemically and physically induced mutants in plants: a Plant Genet. Resour. 2014;12: S74-S78.
CrossRef - Ram H, Soni P, Salvi P, Gandass N, Sharma A, Kaur A, Sharma TR. Insertional mutagenesis approaches and their use in rice for functional genomics. Plants (Basel). 2019; 8:310.
CrossRef - Penna S, Jain SM. Mutant resources and mutagenomics in crop plants. J. Food Agric. 2017; 29: 651-657.
CrossRef - Sikora P, Chawade A, Larsson M, Olsson J, Olsson O. Mutagenesis as a tool in plant genetics, functional genomics, and breeding. J. Plant Genomics 2011;2011:314829.
CrossRef - Rawat N, Schoen A, Singh L, Mahlandt A, Wilson DL, Liu S, Lin G. TILL-D: An Aegilops tauschii TILLING resource for wheat improvement. Plant Sci. 2018; https://doi.org/10.3389/fpls.2018.01665
CrossRef - Viana VE, Pegoraro C, Busanello C, Costa de Oliveira Mutagenesis in rice: the basis for breeding a new super plant. Front. Plant Sci. 2019; https://doi.org/10.3389/fpls.2019.01326
CrossRef - Chawade A, Sikora P, Brautigam M, Larsson M, Vivekanand V, Nakash M, Chen T, Olsson Development and characterization of an Oat TILLING-Population and identification of mutations in lignin and β-glucan biosynthesis genes. BMC Plant Biol. 2010;10:86
CrossRef - Szurman-Zubrzycka ME, Zbieszczyk J, Marzec M, Jelonek J, Chmielewska B, Kurowska MM, Krok M, Daszkowska-Golec A, Guzy-Wrobelska J, Gruszka D, Gajecka M, Gajewska P, Stolarek M, Tylec P, Sega P, Lip S, Kudełko M, Lorek M, Gorniak-Walas M, Malolepszy A, Podsiadlo N, Szyrajew KP, Keisa A, Mbambo Z, Todorowska E, Gaj M, Nita Z, Orlowska-Job W, Maluszynski M, Szarejko I. HorTILLUS—A rich and renewable source of induced mutations for forward/reverse genetics and pre-breeding programs in barley (Hordeum vulgare). Front. Plant Sci. 2018; 9: 216.
CrossRef - Kurowska M, Daszkowska-Golec A, Gruszka D, Szurman M, Szarejko I, Maluszynski M. TILLING – A shortcut in functional genomics.” J App Genet. 2011;52: 371-390.
CrossRef - Mahajan R, Tabia S, Raina G, Mangotra N. Assessment of genetic diversity of non-basmati rice of Jammu and Kashmir using microsatellite markers. J. Cereals Oilseeds.2012; 3: 21-27.
CrossRef - Govindaraj M, Vetriventhan M, Srinivasan M. Importance of genetic diversity assessment in crop plants and its recent advances: An overview of its analytical perspectives. Genet. Res. Int. 2015; 2015:431487.
CrossRef - Moose SP, Mumm RH. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 2008;147: 969-977.
CrossRef - Mahajan R, Javed A, Kapoor N. Characterization of genetic diversity of wild pomegranate collected from Himachal Pradesh, India. Plant Sci. 2018;7:2042-2046.
CrossRef - Fuchs M. Pyramiding resistance-conferring gene sequences in crops. Virol. 2017;26:36-42.
CrossRef - Bhandari HR, Bhanu AN, Srivastava K, Singh MN, Shreya, Hemantaranjan A. Assessment of genetic diversity in crop plants – An overview. Plant Agric. Res. 2017; 7:255.
CrossRef - Bhatia D, Sharma R, Vikal Y, Mangat GS, Mahajan R, Sharma N, Lore JS, Singh N, Bharaj TS, Singh K. Marker-assisted development of bacterial blight resistant, dwarf, and high yielding versions of two traditional basmati rice cultivars. Crop Sci. 2011;51:759-7.
CrossRef - Das G, Patra JK, Baek Insight into MAS: A molecular tool for development of stress resistant and quality of rice through gene stacking. Front. Plant Sci. 2017;8:1321.
CrossRef - Cobb JN, Biswas PS, Platten JD. Back to the future: Revisiting MAS as a tool for modern plant breeding. Appl. Genet. 2019;132:647–667.
CrossRef - Rasheed A, Xu X. From markers to genome-based breeding in wheat Appl. Genet. 2019; DOI: 10.1007/s00122-019-03286-4
CrossRef - Sakiyama NS, Ramos HCC, Caixeta ET, Pereira MG. Plant breeding with marker-assisted selection in Brazil. Crop Breed. Biotechnol. 2014;14:54-60.
CrossRef - Mba C, Guimaraes EP, Ghosh K. Re-Orienting crop improvement for the changing climatic conditions of the 21st century. Food Secur. 2012; 1: 7.
CrossRef - Fuchs M. Pyramiding resistance-conferring gene sequences in crops. Opin. Virol. 2017;26:36-42.
CrossRef - Liu Y, Yuan G, Si H, Sun Y, Jiang Z, Liu D, Jiang C, Pan X, Yang J, Luo Z, Zhang J, Ren M, Pan Y, Sun K, Meng H, Wen L, Xiao Z, Feng Q, Yang A, Cheng L. Identification of QTLs associated with agronomic traits in tobacco via a biparental population and an eight-way MAGIC population. Plant Sci. 2022; https://doi.org/10.3389/fpls.2022.878267.
CrossRef - Benson J, Brown-Guedira G, Murphya JP, Snelle C. Population structure, linkage disequilibrium, and genetic diversity in soft winter wheat enriched for Fusarium head blight resistance. Plant Genome. 2011; 5:71-80.
CrossRef - Bennett R. A, Se´guin-Swartz G, Rahman H. Broadening genetic diversity in canola: towards the development of canola quality. Brassica oleracea. Crop Sci. 2012; 52:2030-2039.
CrossRef - Glenn KC, Alsop B, Bell E, Goley M, Jenkinson J, Liu B, Martin C, Parrott W, Souder C, Sparks O, Urquhart W, Ward JM, Vicini JL. Bringing new plant varieties to market: plant breeding and selection practices advance beneficial characteristics while minimizing unintended changes. Crop Sci. 2017;57:1-16.
CrossRef - Sun X, Li X, Lu Y, Wang S, Zhang X, Zhang K, Su X, Liu M, Feng D, Luo S, Gu A, Fu Y, Chen X, Xuan S, Wang Y, Xu D, Chen S, Ma W, Shen S, Cheng F, Zhao J. Construction of a high-density mutant population of Chinese cabbage facilitates the genetic dissection of agronomic traits. Plant; 2022;15:913-924.
CrossRef - Mahmood T, Mustafa HSB, Aftab M, Ali Super Canola: Newly developed high yielding, lodging and drought tolerant double zero cultivar of rapeseed (Brassica napus L.). Genet. Mol. Res. 2019;18:22.
- Zhang H, Mittal N, Leamy LJ, Barazani O, Song BH. Back into the wild-Apply untapped genetic diversity of wild relatives for crop improvement. Evol. Appl. 2016;10: 5-24.
CrossRef - Bohra A, Kilian B, Sivasankar S, Caccamo M, Mba C, Mc Couch S, Varshney R. Reap the crop wild relatives for breeding future crops. Trends Biotech. 2022;40:412-431.
CrossRef - Warburton ML, Rauf L, Marek M, Hussain O, Ogunola Gonzalez JJS. The use of crop wild relatives in maize and sunflower breeding. Crop Sci. 2017;57:1227-1240.
CrossRef - Jacob J, Sujatha M, and Varaprasad SK. Screening of cultivated and wild Helianthus species reveals herbicide tolerance in wild sunflowers and allelic variation at Ahasl1 (Acetohydroxyacid Synthase 1 Large Subunit) locus. Plant Genet. Res. 2016;1:1-9.
CrossRef - Cheema KK, Grewal NK, Vikal Y, Sharma R, Lore JS, Das A , Bhatia D, Mahajan R, Gupta V, Bharaj T S, Singh K. A novel bacterial blight resistance gene from Oryza nivara mapped to 38 kb region on chromosome 4L and transferred to Oryza sativa” Genet Res (Camb). 2009;90:397-407.
CrossRef - Kumari N, Mahajan R, Gupta V, Bhatia D, Gill BK, Komal R, Lore JS, Mangat GS, Singh K. High-resolution genetic mapping of a novel bacterial blight resistance gene Xa-45(T) identified from Oryza glaberrima and transferred to Oryza sativa. Theoretical and Applied Genetics. 2020;133:689-705.
CrossRef - Diepenbrock CH, Gore MA. Closing the divide between human nutrition and plant breeding. Crop Sci. 2015;55:1437-1448.
CrossRef - Van Der Straeten D, Bhullar NK, De Steur H, Gruissem W, Bouis HE, et al.Multiplying the efficiency and impact of biofortification through metabolic engineering. Commun. 2020; 11:5203.
CrossRef - Ahsin M, Hussain S, Rengel Z, Amir M. Zinc status and its requirement by rural adults consuming wheat from control or zinc-treated fields. Environ. Geochem. Health. 2020;42:1877-1892.
CrossRef - Singh U, Praharaj CS, Chaturvedi SK, Bohra A. Biofortification: introduction, approaches, limitations, and challenges. In: Singh U., Praharaj C., Singh S., Singh N. (eds) Biofortification of Food Crops. Springer, New Delhi 2016;DOI: 1007/978-81-322-2716-8
CrossRef - Boonyaves K, Wu TY, Gruissem W, Bhullar NK. Enhanced grain iron levels in rice expressing an iron regulated metal transporter , Nicotinamine Synthase and Ferritin ene Cassette. Plant Sci. 2017; 8:130.
CrossRef - Beasley JT, Bonneau JP, Sánchez-Palacios JT, Moreno-Moyano LT, Callahan DL, Tako E, Glahn RP, Lombi E, Johnson AAT. Metabolic engineering of bread wheat improves grain iron concentration and bioavailability. Plant Biotech. J. 2019; DOI: https://doi.org/10.1111/pbi.13074.
CrossRef - Gunaratna NS, Moges D, De Groote H. Biofortified maize can improve quality protein intakes among young children in Southern Ethiopia. 2019; 11:192.
CrossRef - Vinoth A, Ravindhran R. Biofortification in millets: A sustainable approach for nutritional security. Plant Sci. 2017; 8:29.
CrossRef - Joshi R, Karan R, Singla-Pareek SL, Pareek A. Ectopic Expression Of Pokkali Phosphoglycerate Kinase-2 (Ospgk2-P) improves yield in tobacco plants under salinity stress. Plant Cell Report.2016; 35:27–41.
CrossRef - Hunter MC, Smith RG, Schipanski ME, Atwood LW, Mortensen Agriculture in 2050: Recalibrating targets for sustainable intensification. BioScience. 2017;67:386–391.
CrossRef - Hong Y, Zhang H, Huang L, Dayong L, Song Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front.Plant Sci. 2016;7: 4
CrossRef - Tao JJ, Wei W, Pan WJ, Lu L, Li QT, Ma JB, Zhang WK, Ma B, Chen SY, Zhang JS. An Alfin-Like gene from Atriplex Hortensis enhances salt and drought tolerance and abscisic acid response in transgenic Sci. Rep. 2018;8: 2707.
CrossRef - Borisjuk N, Kishchenko O, Eliby S, Schramm C, Anderson P, Jatayev S, Kurishbayev A, Shavruk Y. Genetic modification for wheat improvement: from transgenesis to genome editing. BioMed Res. Int. 2019; 2019:6216304.
CrossRef - Qu B, Xe X, Wang J, Zhao Y, Teng W, Shao A, Zhao X, Ma W, Wang J, Li B, Li Z, Tong YA. Wheat CCAAT Box-Binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 2015;167: 411–423.
CrossRef - Kondou Y., Higuchi M., Matsui M. High-Throughput characterization of plant gene functions by using gain-of-function technology. Annu. Rev. Plant Biol. 2010;61:373-393.
CrossRef - Abe K., Ichikawa H. Gene overexpression resources in cereals for functional genomics and discovery of useful genes. Plant Sci.2016;7:1359.
CrossRef - Yoshihiko O, Keiichi Exploring genetic diversity in plants using high-throughput sequencing techniques. Curr. Genom. 2016;17: 358-367.
CrossRef - Singh UM, Chandra M, Shankhdhar SC, Kumar A. Transcriptome wide identification and validation of calcium sensor gene family in the developing spikes of finger millet genotypes for elucidating its role in grain calcium accumulation. PLoS One. 2014;9: e103963.
CrossRef - Rahman H, Jagadeeshselvam N, Valarmathi R, Sachin B, Sasikala R, Senthil N, Sudhakar D, Robin S, Muthurajan R. Transcriptome analysis of salinity responsiveness in contrasting genotypes of Finger Millet (Eleusine Coracana) through RNA-sequencing. Plant Molecular Biology. 2014; 85:485-503.
CrossRef - Jaganathan D, Bohra A, Thudi M, Varshney R. Fine mapping and gene cloning in the post-NGS era: advances and prospects. Appl. Gene.2020;133:1791-1810.
CrossRef - Unamba CIN, Nag A, Sharma Next Generation Sequencing Technologies: The doorway to the unexplored genomics of non-model plants. Front. Plant Sci. 2015;6: 1074.
CrossRef - Manghwar H, Lindsey K, Zhang X, Jin S. CRISPR/Cas system: Recent advances and future prospects for genome editing. Trends Plant Sci. 2019;24:1102–1125
CrossRef - Hua K, Zhang J, Botella JR, Ma C, Kong F, Liu B, Zhu JK. Perspectives on the application of genome-editing technologies in crop breeding. Plant. 2019;12:1047-1059.
CrossRef - Carroll D. Genome engineering with zinc-finger nucleases. 2011;188:773-782.
CrossRef - Joung JK, Sander JD. TALENs: A widely applicable technology for targeted genome editing. Rev. Mol. Cell Biol. 2013;14: 49-55.
CrossRef - Zhang Y, Massel K, Godwin ID. Applications and potential of genome editing in crop improvement. Genome Biol. 2018;19: 210.
CrossRef - Jaganathan D, Ramasamy K, Sellamuthu G, Jayabalan S, Venkataraman G. CRISPR for crop improvement: An update review. Plant Sci. 2018; 9: 985.
CrossRef - Khan ZA, Kumar R, Dasgupta I. CRISPR/Cas-mediated resistance against viruses in plants. Int J Mol Sci. 2022; 23: 2303.
CrossRef - Mishra R, Zheng W, Joshi RK, Kaijun Genome editing strategies towards enhancement of rice disease resistance. Rice Sci. 2021;28:133-145.
CrossRef - Zhou J, Peng Z, Long D, Sosso D, Liu B, Eom JS, Huang S, Liu S, Vera Cruz C, Frommer WB, White FF, Yang B. Gene targeting by the TAL Effector Pthxo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015;82:632–643.
CrossRef - Wang W, Pan Q, He F, Akhunova A, Chao S, Trick H, Akhunov E. Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 2018;1: 65-74.
CrossRef - Zhu J, Song N, Sun S, Yang W, Zhao H, Song W, Lai J. Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. Genet. Genom. 2016; 43: 25-36.
CrossRef - Young J, Zastrow-Hayes G, Deschamps S, et al. CRISPR-Cas9 editing in maize: systematic evaluation of off-target activity and its relevance in crop improvement. Rep. 2019; 9:6729 .
CrossRef - Li W, Chern M, Yin J, Wang J, Chen X. Recent advances in broad-spectrum resistance to the rice blast disease. Opin. Plant Biol. 2019; 50:114-120.
CrossRef - Ning X, Yunyu W, Aihong L. Strategy for use of rice blast resistance genes in rice molecular breeding. Rice Sci. 2020;27:263-27.
CrossRef - Hour AL, Hsieh WH, Chang SH, Wu YP, Chin HS, Lin YR. Genetic diversity of landraces and improved varieties of rice (Oryza sativa) in Taiwan. Rice. 2020; 13:82.
CrossRef - Hassan DA, Hama-Ali EO. Evaluation of gene flow and genetic diversity in rice accessions across Kurdistan region-Iraq using SSR markers. Mol Biol Rep. 2022; 49:1007–1016.
CrossRef - Boonyaves K, Wu T, Wu R, Gruissem W, Bhullar NK. Enhanced grain iron levels in rice expressing an iron regulated metal transporter, nicotinamine synthase and ferritin gene cassette. Plant Sci. 2017; 8:130.
CrossRef - Gaikwad KB, Singh N, Bhatia D, Kaur R, Bains NS, Bharaj TS, Singh K. Yield-enhancing heterotic QTL transferred from wild species to cultivated rice Oryza sativa PLoS ONE. 2014; 9:e96939.
CrossRef - Pradhan SK, Nayak DK, Mohanty S, Behera L, Barik SR, Pandit E, Lenka S, Anandan A. Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice. 2015;8:19.
CrossRef - Yang RC, Peng FY, Hu Z. Inferring defense-related gene families in Arabidopsis and wheat. BMC Genom. 2017;18: 980.
CrossRef - Babu P, Baranwal DK, Harikrishna Pal D, Bharti H. Application of genomics tools in wheat breeding to attain durable rust resistance. Plant Sci. 2020; https://doi.org/10.3389/fpls.2020.567147
CrossRef - Wani SH, Gaikwad K, Razzaq A, Samantara K, Kumar M, Govindan V. Improving zinc and iron biofortification in wheat through genomics approaches. Mol. Biol. Rep. 2022; 49:8007–8023.
CrossRef - Yang T, Yao S, Hao L, Zhao Y, Lu W, Xiao K. Wheat Bhlh-Type transcription factor gene Tabhlh1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-Associated pathway. Plant Cell Rep. 2016; 35:2309-2323.
CrossRef - Lohithaswa HC, Jyothi K, Kumar KRS, Hittalmani S. Identification and introgression of QTLs implicated in resistance to sorghum downy mildew (Peronosclerospora sorghi (Weston And Uppal) C. G. Shaw) in maize through marker-assisted selection. Genet. 2015; 94:741-748.
CrossRef - Zunjare RU, Hossain F, Muthusamy V, Baveja A, Chauhan HS, et al. Development of biofortified maize hybrids through marker-assisted stacking of β-carotene hydroxylase, lycopene-ε-cyclase and opaque2 genes. Plant Sci. 2018;9:178.
CrossRef - Hussain M, Rauf S, Riaz MA, Al-Khayri JM, Monneveux P. Determination of drought tolerance related traits in Helianthus argophyllus, Helianthus annuus, and their hybrids. Sci. 2017; 67:257–267.
CrossRef - Killi D, Bussotti F, Raschi A, Haworth M. Adaptation to high temperature mitigates the impact of water deficit during combined heat and drought stress in C3 sunflower and C4 maize varieties with contrasting drought tolerance. Plant. 2017; 159:130-147.
CrossRef - Singh M, Nara U. Genetic insights in pearl millet breeding in the genomic era: challenges and prospects. Plant Biotechnol. Rep.2022; https://doi.org/10.1007/s11816-022-00767-9.
CrossRef - Yao Y, You Q, Duan G, Ren J, Chu S, Zhao J, Li X, Zhou X, Jiao Y. Quantitative trait loci analysis of seed oil content and composition of wild and cultivated soybean. BMC Plant Biol. 2020; 20:51.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





