How to Cite | Publication History | PlumX Article Matrix
Akhilesh Kumar* , Vijay Kumar
, Vijay Kumar , Anjali Uniyal
, Anjali Uniyal , Sanjay Gupta
, Sanjay Gupta and Vivek Kumar
and Vivek Kumar
Himalayan School of Biosciences, Swami Rama Himalayan University Dehradun, Uttarakhand-248140, India.
Corresponding Author E-mail:akhibiochemist06@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3123
ABSTRACT: For the DNA-based study of plant species, one of the important steps is to obtain high-quality DNA. However, this is problematic when the species contains a lot of polyphenols and polysaccharides. The polysaccharides and polyphenols interfere with the activity of the Taq polymerase enzyme during the PCR reaction thereby affecting the quality of the DNA. Therefore, a method for DNA extraction from Cyclanthera pedata has been developed. The current study reveals a CTAB-based approach that is quick, dependable, and economical and is specifically designed for obtaining DNA from the Cyclanthera genus. These plant species are abundant in secondary metabolites and polysaccharides, which makes it difficult to extract DNA effectively and with a high yield. The present protocol also excludes the use of expensive liquid nitrogen, which makes it cost-friendly as well. High salt concentration (1.5 M) and 2% polyvinylpyrrolidone were used in the DNA extraction buffer to prevent the solubility of polysaccharides and polyphenols in DNA extract. In addition to these substances, protein-like various enzymes were precipitated by ammonium acetate and removed by centrifugation during the isolation process. The quality of the isolated DNA was assessed using agarose gel electrophoresis (0.8%) and quantified using an A260/A280 ratio ranging from 1.7 to 1.9, absorbance ratio >2,which indicates the extract was free of proteins, polysaccharides, and polyphenols. The extracted genomic DNA was amplified by the ISSR primer (UBC-825) and clear banding pattern were observed. This standardized protocol provides pure and high quality genomic DNA without expensive liquid nitrogen or toxic phenolic compounds. It is also suitable for routine molecular biology assays such as RAPD, SSR, restriction digestion, southern blot, and cloning techniques.
KEYWORDS: Cyclanthera pedata; CTAB; Genomic DNA; ISSR; Liquid nitrogen; PVP
Download this article as:| Copy the following to cite this article: Kumar A, Kumar V, Uniyal A, Gupta S, Kumar V. An Effective and Low-Cost Method for Genomic DNA Extraction from Cyclanthera Pedata Species (A Nutraceutical Plant) without Liquid Nitrogen for Inter Simple Sequence Repeat Analyses. Biosci Biotech Res Asia 2023;20(2). |
| Copy the following to cite this URL: Kumar A, Kumar V, Uniyal A, Gupta S, Kumar V. An Effective and Low-Cost Method for Genomic DNA Extraction from Cyclanthera Pedata Species (A Nutraceutical Plant) without Liquid Nitrogen for Inter Simple Sequence Repeat Analyses. Biosci Biotech Res Asia 2023;20(2). Available from: https://bit.ly/3piCxQt |
Introduction
C. pedata is a herbaceous plant, basically a creeper, grown for its edible fruit, which is mostly used as a vegetable. Because of its bitterless taste, it is locally known as pahari karela/meetha karela in Uttarakhand, India. C. pedata is a vine that may grow up to 12 meters in length, with thin stems, and leaves that can grow up to 24 cm long and have either a palmate or pedate shape. Its fruits regulate the metabolism of fats and lower blood cholesterol because they are 93% water by weight. It is utilized in diets for weight loss due to its low calorie content. The fruit of this plant is specifically rich in salts and minerals like iron, zinc, phosphorus, selenium, magnesium, calcium etc. as well as several classes of polyphenolic compounds like flavonoids and phenolic acids. It also contains carbohydrates, pectin, albuminoidal matter, lipid substances, proteins, vitamins C1&2. The concentration of these phytochemicals may depend on many factors, including climatic conditions3. Due to its nutraceutical and medicinal value, it is necessary to know its genetic architecture to evaluate its genetic makeup. DNA-based molecular markers thus play an important role in genetic studies such as fingerprinting of genotypes and analysis of genetic diversity.
For the isolation of good quality genomic DNA, the traditional molecular studies methods employ the use of juvenile leaf tissues as the basic source. The pre-existing methods for the isolation of genomic DNA suggested by previous authors are incompetent for extracting DNA from plant tissues without the use of liquid nitrogen4&5. Therefore, it was necessary to optimize and standardize the protocol for DNA extraction from fruit pulp in order to obtain high-quality DNA at high concentrations suitable for polymerase chain reaction (PCR) applications. In this present research, a modified non-liquid nitrogen-based DNA extraction protocol has been reported. For molecular-based research, the protocol might provide relatively high-quality DNA.
Materials and Methods
Plant material
For DNA extraction, a young fruit sample of C. pedata was brought from the local market of Dehradun. A minimum of 10 individuals were taken and labeled in sterile plastic bags separately. After their arrival at the laboratory, the samples were transferred to a -20°C freezer to maintain the quality for further analysis.
Solutions and Reagents
The solutions and reagents used in DNA isolation were:
CTAB (Cеtyl-trimеthylammoniumBromidе) Buffer pH-8.0 (100 mM Tris-HCl, 20 mM EDTA (Ethylenediaminetetraacеtic acid), 1.5 M NaCl, 2% CTAB, 2% polyvinylpyrollidonе (PVP), and 5 mM Ascorbic acid), 0.3% β-mercaptoеthanol, 5 M Amoniumacetatе solution, Chloroform: Isoamyl alcohol (24:1), 76% ethanol and chilled 96% isopropanol, TE buffer pH- 8.0 (Tris-HCl 10 mM, EDTA 1mM), Wash buffer: 998 μl of 96% Ethanol and 2 μl of ammonium acetate.
Genomic DNA Isolation
The total genomic DNA was extracted from young fruit tissue of pedata (Figure 1) by used a slightly modified protocol of Doyle and Doyle4.
The sliced and chopped fruit pulp (1.5gm) was grounded to a fine liquid used a mortar pestle with 3 ml of extraction buffer.
A 2 ml eppendorf tube was filled with 1 ml of ground sample and 3 μl of β-mercaptoethanol. The suspension was vortexed for 15 seconds and incubated at 60°C for 30 minutes.
Allow the samples to cool to room temperature before adding an equal volume of chloroform and isoamyl alcohol in a 24:1 ratio, followed by inversion mixing in eppendorf tube. Centrifuge at 4 °C for 6 minutes at 10,000 rpm.
A new eppendorf tube was used to pipette out the upper aqueous phase (1.5 ml).
For the purpose of precipitating genomic DNA, 500 µl of chilled isopropanol was added to the supernatant, thoroughly mixed, and incubated overnight at -20 °C.
To obtain the pellet, centrifuge at 4 °C for 10 minutes at 10,000 rpm.
The pellet was washed with 998 µl of 96% ethanol and 2 µl of ammonium acetate after centrifugation.
The pellet was subsequently washed once more with 500 µl of 76% alcohol, centrifuged at 10,000 rpm for 6 minutes, vacuum dried, and re-suspended in 100 µl of TE buffer, where it was kept at 4 °C until it was needed.
Qualitative and Quantitative DNA Analysis
Using a UV-VIS spectrophotometer (Shimadzu), the yield and purity of the genomic DNA that was extracted were evaluated. Analyzing the absorbance ratio A260/A280 allowed for an evaluation of DNA purity. All of the extracted DNA samples were run on an agarose gel at a concentration of 0.8% to facilitate visualization, which was done using the GEL Documentation System (Analytic Jena).
 |
Figure 1: Pictorial representation of genomic DNA extraction steps (A) Individually sliced C. pedata fruit tissue ready to crush in CTAB buffer. (B) Fine liquid tissue ready to centrifuge. |
Molecular analysis through ISSR Primers
The ISSR primer (the University of British Columbia, UBC-825 Sequence: 5’ACACACACACACACACT-3’ was used for the amplification6. The PCR amplification was carried out in a 20µl reaction mixture (Table 1). The amplification cycle comprise of one cycle of denaturation at 94°C for 5 min, 40 cycles each of 94°C for 30 seconds, 52.6°C (annealing) for 30 seconds, and 72°C (extension) for 1 minute, followed by a final extension at 72°C for 8 minutes. The reaction mixture was kept on hold at 4°C till taken for storage. The PCR products were visualised on a 1% agarose gel, and bands were observed in the gel documentation system (Figure 2).
Table 1: Concentration of PCR reaction mixture for ISSR.
|
S. No |
Stock |
Reaction |
Final concentration |
|
1 |
Taq Buffer (10X) |
2.00µl |
1X |
|
2 |
MgCl2 (25mM) |
1.40µl |
1.75mM |
|
3 |
dNTPs (2.5mM) |
1.60µl |
0.2mM |
|
4 |
Primer (20µM) |
0.40µl |
0.4µM |
|
5 |
Taq DNA Polymerase (5U) |
0.12µl |
0.03U |
|
6 |
Template DNA |
1.00µl |
15ng/µl |
|
7 |
Deionised Water |
13.48 µl |
– |
|
8 |
Total volume |
20.00µl |
– |
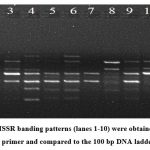 |
Figure 2: ISSR banding patterns (lanes 1-10) were obtained with the UBC-825 primer and compared to the 100 bp DNA ladder lane L. |
Results and Discussions
From the ten (10) individual C. pedata fruit samples, high-quality genomic DNA was effectively isolated. The A260/A280 ratio was between 1.7 and 1.9, which is within the ideal sample range, and the extracted DNA had a high level of purity and it was closer to the findings of Sambrook et al., 1989 and Singh et al., 20217&8. Proteins and fatty acids were the primary components of C. pedata fruits. When centrifuging, fatty acids can be easily differentiated from the aqueous phase due to their lower density and non-polar properties. In conventional DNA extraction protocols, phenol and chloroform together are frequently used for protein removal. Only a short centrifugation spin is required to separate DNA from all the other contaminants in our present protocol. Most of the protein part contaminants were removed in the insoluble precipitate. The extraction buffer contained 1.5 M NaCl, which facilitates the removal of the polysaccharides by increasing their solubility in ethanol and existing results revealed similar to Fung et al., 1992 and Heikrujam et al., 2020 9&10. PVP (2%) was added to the extraction buffer to remove polyphenols from the fruit tissues and this result was closer to the findings of Maliyakal 199211. To protect DNA from oxidation and degradation, 5 mM ascorbic acid and 0.3% β-mercaptoethanol were used and the result was justified by previous findings of Kumar et al., 201512. The high quality of these 10 extracted DNA samples was confirmed by the ISSR molecular markers’ positive amplification results. The results show the protocol’s value for molecular marker research using the method of DNA extraction from C. pedata. For molecular research on plants that contain huge amount of polyphenols and polysaccharides, it is incredibly helpful.
Conclusions
The objective of the study was to isolate high quality genomic DNA for PCR amplification from young fruit pulp of C. pedata. The protocols, developed are highly efficient, simple, and cost-effective for extracting genomic DNA even from plant tissues which contains high polysaccharides and polyphenols.
Acknowledgement
Authors are thankful to the Himalayan school of Biosciences, Swami Rama Himalayan University, for providing the proper infrastructure and lab facility to carry out this research work.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Funding Sources
There was no specific grant for this research from any funding organization in the public, private, or nonprofit sectors.
References:
- Janick, J., & Paull, R.E. The Encyclopedia of Fruit and Nuts (1st Edition), CABI; 2008. pp 299. ISBN9780851996387.
CrossRef - Sukorno F.I., Islam S., Kabir A.L., Cruz C.V., Zaman S., Ishaku G.A. Phytochemicals are natural resources of food supplement for happier people. Horticulture International Journal. (2019); 3(6):300‒305.DOI: 10.15406/hij.2019.03.00145
CrossRef - Li H, Tsao R, Deng Z. Factors affecting the antioxidant potential and health benefits of plant foods. J. Plant Sci. 2012; 92: 1101-1111.
CrossRef - Doyle J.J., & Doyle J.L. Isolation of plant DNA from fresh tissue. Focus. 1990; 12: 13–15.
CrossRef - Stange C, Prehn D, Arce P. Isoaltion of Pinusradiata genomic DNA suitable for RAPD analysis. Plant Mol. Biol. Rep. 1998; 16: 1-8.
CrossRef - Hong Y.P., Kwon H.Y., Kim I.S. ISSR markеrs reveald in consistеnt phylogеographic pattеrns among populations of Japanesе red pinеs in Korеa. Silvaе Genеt. 2007; 43:167-176.
- Sambrook J., Fritsch E.F., Maniats T. Molеcular Cloning: a Laboratory Manual. Cold Spring Harbor Laboratory Prеss. New York; 1989
- Singh R, Yadav S.K., Roy S, Thapliyal M, Ginwal H.S., & Barthwal S. High Quality DNA extraction protocol for cotyledonary and mature leaves of Rhododenderon aroboreum Sm. Indian Forestor. 2021; 147(11) 1044-1050 DOI: 10.36808/if/2021/v147i11/157680
CrossRef - Fung G, Hammar S, Rebecca R. A quick and in expensive method for removing polysaccharides from plant genomic DNA. Biotechnology. 1992; 13:52-56.
- Heikrujam J, Kishor R, Mazumder P.B. The chemistry behind plant DNA isolation protocols. Biochemical Analysis Tools–Methods for Bio-Molecules Studies. IntechOpen. 2020; pp 206 DOI: 10.5772/intechopen.82530
CrossRef - Maliyakal E. J. An efficient method for isolation of RNA and DNA from plants containing polyphenolics. Acids Res. 1992; 20: 2381.
CrossRef - Kumar A, Barthwal S, Ginwal H.S. Genomic DNA isolation from old needles of Cedrus deodara (Roxb.) G. Don and amplification of nuclear microsatellite markers. Indian Journal of Forestry. 2015; 38 (3) 215-217
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





