How to Cite | Publication History | PlumX Article Matrix
Puja Banduji Paunfase 1 , Samynathan Ramkumar2
, Samynathan Ramkumar2 , Marappan Ganesan2
, Marappan Ganesan2 and Veeraraghavan Usha1*
and Veeraraghavan Usha1*
1Department of Biotechnology (PG), PSGR Krishnammal College for Women, Coimbatore, India.
2Alchem Diagnostics, 3/45 Theethipalayam, Coimbatore - 641010, India
Corresponding Author Email:ushaveera13@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3159
ABSTRACT: Dental caries, a highly prevalent infectious disease in humans is caused by the bacterial plaque that coats the teeth surface and is a serious public health concern. Recently, the formation of dental plaque has been associated with the occurrence of various other systemic diseases, Alzheimer's disease, Cardiovascular diseases, Rheumatoid Arthritis, Respiratory diseases, Bacteremia and Cancer. Despite the fact that both Streptococcus mutans and Streptococcus sobrinus are the major etiologic agents of dental caries, S. mutans is more prevalent than S. sobrinus in dental plaque. Early detection of S. mutans and S. sobrinus was carried out from five caries affected dental plaque samples collected from Sri Ramakrishna Dental College and Hospital, Coimbatore, by using semi-quantitative real-time PCR. Specific primers for gtfB and gtfI genes of S. mutans and S. sobrinus respectively were used for the quantification of cariogenic bacteria in the given dental plaque samples. The Biopro Oral Microbiome transport media was prepared to carry dental plaque samples from the hospital to the laboratory. Genomic DNA extraction was done by employing magnetic beads and spin columns provided in the Biopro DNA isolation kit. Various biochemical tests were performed on the bacterial cultures isolated from dental plaque.
KEYWORDS: Dental caries; Dental plaque; Polymerase Chain Reaction; Streptococcus mutans; Streptococcus sobrinus
Download this article as:| Copy the following to cite this article: Paunfase P. B, Ramkumar S, Ganesan M, Usha V. Detection of Streptococcus mutans and Streptococcus sobrinus in Human Dental Plaque Samples by Using Semi-Quantitative Real-Time Polymerase Chain Reaction. Biosci Biotech Res Asia 2023;20(3). |
| Copy the following to cite this URL: Paunfase P. B, Ramkumar S, Ganesan M, Usha V. Detection of Streptococcus mutans and Streptococcus sobrinus in Human Dental Plaque Samples by Using Semi-Quantitative Real-Time Polymerase Chain Reaction. Biosci Biotech Res Asia 2023;20(3). Available from: https://bit.ly/3RKsnUm |
Introduction
The “Microbiome” found in a healthy human body comprises of 3.8 x 1013 bacterial cells and other microorganisms1. One of the body’s most complex microbiomes is found in the oral cavity, and oral bacteria plays a major role in the development and occurrence of Dental Caries (tooth decay)2. Dental caries is a commonly occurring chronic illness that develops as a result of the acidic demineralization of tooth tissues brought about by bacterial fermentation of dietary carbohydrates3. Around the world, 3.5 billion people suffer from oral diseases, 2 billion people are thought to have Dental Caries, and 514 million children have primary tooth decay4.
Dental plaque consists of a physically and functionally organized, species-rich microbial community5. Approximately 80% of the microorganisms of the biofilm is dominated by Streptococcus which has a role in the initial colonization process, followed by Actinomyces and other bacteria6. During high availability of fermentation substrates, Streptococcus species has the ability to ferment carbohydrates into acetic, formic and lactic acid which in turn reduces pH values near five7. Many strong acid-producing and acid-tolerant organisms, including S. mutans, interact with other biofilm residents and are the principal causal agents of dental caries8,9. S. mutans, a gram-positive facultative anaerobic bacteria, is typically found in biofilms on tooth surfaces, or in dental plaque10.
The two Steptococcus species, S. mutans and S. sobrinus typically co-exist in a person independently, with S. mutans being more common in areas around caries and having a close association with the disease condition11. When cultivated on plates using Mitis salivarius agar, a selective medium for S. mutans, S. mutans colony morphology is rough, whereas S. sobrinus is smooth12. Another important characteristic that explains why S. mutans is the main causative agent of dental caries is due to its ability to adapt to abrupt and significant environmental changes inside the tooth plaque13. S. mutans can withstand frequent and quick environmental changes such as nutrition availability, aerobic to anaerobic transitions, and pH shifts because it uses carbohydrates to create biofilm on tooth surfaces14.
Despite the fact that caries is a multi-microbial illness, targeting S. mutans specifically in dental biofilms is thought to be an effective strategy for its prevention13. This is primarily due to the fact that S. mutans GTFs (glucosyltransferases), particularly water-insoluble glucan-synthesizing GTFs, are associated with the bacteria’s cariogenicity10. The water-insoluble glucans (also called mutans), are rich in alpha (1→3) linkages produced by gtfB gene of S. mutans, are essential for the development of a stable biofilm matrix13,15. This matrix in turn promotes bacterial colonization on the tooth surface and also acts as a diffusion barrier, maintaining the acidic environment necessary for cariogenic bacteria to flourish16. Both the S. mutans gtfB gene and the S. sobrinus gtfI gene encode for glucosyltransferase enzyme10.
A semi-quantitative real-time PCR reaction was performed to detect 16S rRNA in the genomic DNA purified from dental plaque samples in order to confirm the presence of bacteria in the dental plaque samples. In the current study the presence of S. mutans and S. sobrinus was confirmed in the subgingival plaque samples by using species specific primers of glucosyltransferase gene and real-time polymerase chain reaction for detection.
Materials and Methods
Collection of sample
The clinical dental plaque samples were aseptically collected from the subgingival region of five randomly selected patients visiting Sri Ramakrishna Dental College & Hospital, Coimbatore. The dental plaque samples were scraped from the tooth surface of the patients using a dental explorer, collected in sterile swabs attached to toothpicks and were immediately dipped in Oral Microbiome transport media. Collection tubes were sealed with parafilm to prevent contamination during transportation and were stored at 4℃.
Preparation of Oral Microbiome transport medium
The Biopro Oral microbiome transport medium (50 ml) was prepared at Alchem Diagnostics, Coimbatore for carrying dental plaque samples. This Oral microbiome transport medium consists of modified Hanks basal salt solution and a buffer to maintain the pH at 7.3 +/- 0.2. The medium also includes protein for stabilisation. It helps store samples for long periods by preserving bacteria when stored frozen in the presence of a cryoprotectant. The Oral microbiome transport medium was sterilized using 0.22 µm PES filter and stored at temperatures between 10 to 30℃.
Preparation of Genomic DNA from plaque sample
The dental plaque samples dissolved in 200µL of oral microbiome transport medium were harvested by centrifugation at 5000 x g for 10 minutes in order to obtain the cell pellet. The genomic DNA was isolated from the cell pellet and supernatant was discarded. Bacterial genomic DNA extraction was carried out from the five dental plaque samples by using Biopro DNA extraction kit. The protocol from the manufacturer’s instruction manual was followed except for the inclusion of an RNase treatment step. The Biopro DNA Extraction kit provides a simple, convenient and reliable protocol for the isolation of high quality DNA using unique buffers. The basic four steps used in the extraction of DNA by using the Biopro DNA extraction kit are: Lysis, Binding, Washing and Elution. The kit was used to isolate DNA either by employing magnetic beads or spin column.
Agarose gel electrophoresis
The concentrations of DNA purified by using Biopro magnetic beads and spin columns were determined by using the Nanodrop Spectrophotometer (Thermo Scientific, USA). The Absorbance at 260 nm of the DNA samples was measured and the concentrations calculated. The Absorbance of the DNA samples were also measured at 280 nm in order to estimate the purity of DNA from the A260/A280 ratio. Agarose gel electrophoresis of the isolated DNA samples confirmed the DNA quality.
Semi-quantitative Real-time Polymerase chain reaction
The purified genomic DNA samples from dental plaques had to be confirmed with 16S rRNA universal primers whether it is of bacterial origin. Using gene specific primers or 16S rRNA universal primers, the reaction mixture in a total volume of 20μl contained 2μl of nuclease free water, 10μl SYBR® green qPCR master mix, 1.5μl each of forward and reverse gene specific primers or 1.5μl each of forward and reverse 16S rRNA universal primer, and 5μl of bacterial genomic DNA. The C1000™ thermocycler with the CFX 96 Real-time System (Biorad, USA) was set for 40 cycles. Each cycle consisted of an enzyme activation step at 95°C for 10 minutes followed by denaturation at 95°C for 15 seconds, annealing and extension at 60°C for 1 minute. The amplification curve was obtained for each gene specific primer or 16S rRNA universal primer at the end of each PCR reaction. A negative control (water blank) was included while performing the PCR. Sterile deionized water was used as a negative control to detect any potential contamination. The sequences of the 16S rRNA universal primer, S. mutans gtfB and S. sobrinus gtfI primers are listed in Table No. 1
Table 1: Primers designed for semi-quantitative RT-PCR reaction. Sequence of 16S rRNA universal primer, S. mutans gtfB and S. sobrinus gtfI gene primers.
|
Target gene |
Primer |
Primer sequence (5’→ 3’) |
Amplicon length (bp) |
References |
|
16S rRNA (Universal primer) |
Forward |
TGGAGCATGTGGTTTAATTCGA
|
160 |
Reference17 |
|
Reverse |
TGCGGGACTTAACCCAACA
|
|||
|
gtfB of S. mutans |
Forward |
AGCCATGCGCAATCACAGGTT
|
415 |
Reference10 |
|
Reverse |
CGCAACGCGAACATCTTGATCAG
|
|||
|
gtfI of S. sobrinus |
Forward |
GAAACCAACCCAACTTTAGCTTGGAT |
319 |
Reference18 |
|
Reverse |
ATGGAGTGATTTTCCATCGGTACTTG |
Culture conditions
The dental plaque samples collected in the Biopro oral microbiome transport media were streaked on sterile Nutrient agar plates. The plates were incubated overnight at 37℃. The microbial colonies grown were examined by Gram’s staining & subjected to various biochemical tests.
Biochemical tests – Catalase test
A loop-full of all the five overnight grown cultures were transferred on five clean, well labelled microscopic glass slides in a sterile environment. After the addition of a drop of 3% Hydrogen peroxide on each glass slide, the slides were observed for the formation of gas bubbles19.
Urease test
The tubes containing Urea agar slants were inoculated with a loopful of overnight grown culture and the tubes were incubated at 37℃ for 48 hours and observed for colour change20.
Voges-Proskauer (VP) test
The VP medium containing tubes were inoculated with a loopful of overnight grown culture and incubated for 24 hours at 37℃, 2 ml of fresh VP broth was added to the test tubes and the incubation at 37℃ was continued for 24 hours. Approximately 6 drops of 5% alpha-naphthol were added to the cultures, mixed well by stirring, followed by the addition of 2 drops of 40% potassium hydroxide. The tubes were observed for the pink red colour formation on the surface of medium by vigorously shaking for 30 minutes21.
Fermentation of Glucose
The phenol red glucose broth was prepared in flasks by dissolving Trypticase, Sodium chloride, and Phenol red in a minimal volume of distilled water, after addition of 0.5% to 1% of glucose the final volume was made upto 100 ml with water. In order to detect gas production Durham or Fermentation tubes were inverted and completely immersed in the broth and the set up was autoclaved for 15 minutes at 121°C. During the sterilization process, the broth would be filled in the inverted fermentation tubes. Each tube was inoculated with 1 drop of an overnight grown culture under sterile conditions. An un-inoculated fermentation tube was used as control. The fermentation tubes were incubated for 18 to 24 hours at 37°C and observed for a visible air bubble which indicated gas production. The appearance of a yellow colour indicated that enough acid was produced due to the fermentation of glucose22
Results
Isolation and purification of bacterial genomic DNA from dental plaque samples
From the five dental plaque samples, bacterial genomic DNA was isolated and purified. The yield and quality of DNA obtained was analysed by using Nanodrop Spectrophotometer (Data not shown). The DNA quantification data showed better quantity of DNA isolated using spin column when compared with DNA isolated by using magnetic beads. Similarly, the quality of spin column purified DNA was better than the quality of DNA purified by using magnetic beads. The amount of genomic DNA recovered from the dental plaque samples was sufficient for performing the semi-quantitative RT-PCR experiment.
Biochemical characteristics of culture
The biochemical tests carried out showed convincing results for all the cultures grown from the five dental plaque samples. Gram staining showed the presence of purple-coloured cocci when observed under a light microscope. Cultured bacteria were shown to be catalase and urease negative. All the sample cultures were found to be positive for the Voges-Proskauer (VP) test and glucose fermentation test as shown in Table No. 2.
Table 2: Biochemical characteristics of cultures grown from five dental plaque samples.
|
Sample |
Gram Staining |
Catalase |
Urease |
VP |
Fermentation of Glucose |
|
1 |
Cocci + |
– |
– |
+ |
+ |
|
2 |
Cocci + |
– |
– |
+ |
+ |
|
3 |
Cocci + |
– |
– |
+ |
+ |
|
4 |
Cocci + |
– |
– |
+ |
+ |
|
5 |
Cocci + |
– |
– |
+ |
+ |
Semi-quantitative Real-time PCR detection of S. mutans and S. sobrinus
The purified genomic DNA was confirmed for the presence of bacterial species by targeting bacterial 16S rRNA gene. Semi-quantitative Real-time PCR was performed using 16S rRNA forward and reverse primers (Figure No. 1). The short oligonucleotide primers complementary to the hypervariable regions of 16S rRNA were specifically designed to detect bacterial species. The qRT-PCR result using 16S rRNA Universal primers with cycle threshold values of less than 20 shown in Figure No. 1 and Table No. 3 confirmed that the extracted DNA was of bacterial origin. In order to specifically detect the two oral streptococcal species (S. mutans and S. sobrinus) using the Semi-quantitative Real-time PCR method, specific primer sets encoding the glucosyltransferase gene (S. mutans gtfB and S. sobrinus gtfI) were employed. S. mutans gtfB gene was detected at cycle threshold values ranging from 31-35 for spin column purified DNA (Figure No. 2a and Table No. 3) whereas, cycle threshold values for DNA purified using magnetic beads for gtfB gene of S. mutans were ranging around 30-31 (Figure No. 2b and Table No. 3). The gtfI gene of S. sobrinus was detected at cycle threshold values of less than 40 for spin column purified DNA (Figure No. 3a and Table No. 3) whereas, cycle threshold values for DNA purified using magnetic beads for gtfI gene of S. sobrinus were ranging around 31-34 (Figure No. 3b and Table No. 3).
Table 3: Cycle threshold (Ct) values obtained for 16S rRNA, S. mutans gtfB and S. sobrinus gtfI gene amplification.
|
Dental plaque sample
|
Cycle threshold (Ct) Values |
||||
|
Target gene: 16S rRNA |
Target gene: gtfB (S. mutans) |
Target gene: gtfI (S. sobrinus) |
|||
|
Spin column purified DNA |
Spin column purified DNA |
Magnetic beads purified DNA |
Spin column purified DNA |
Magnetic beads purified DNA |
|
|
1 |
20.29 |
33.02 |
30.30 |
39.33 |
25.33 |
|
2 |
20.21 |
33.80 |
31.18 |
36.57 |
31.12 |
|
3 |
20.23 |
34.31 |
30.01 |
38.64 |
32.20 |
|
4 |
20.52 |
34.90 |
31.00 |
35.44 |
34.11 |
|
5 |
20.20 |
31 |
30.55 |
36.38 |
33.11 |
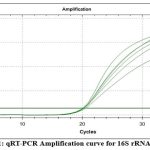 |
Figure 1: qRT-PCR Amplification curve for 16S rRNA gene
|
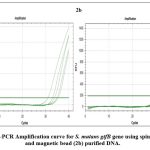 |
Figure 2: qRT-PCR Amplification curve for S. mutans gtfB gene using spin column (2a) and magnetic bead (2b) purified DNA.
|
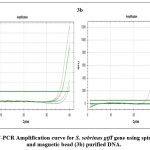 |
Figure 3: qRT-PCR Amplification curve for S. sobrinus gtfI gene using spin column (3a) and magnetic bead (3b) purified DNA.
|
Discussion
The most widespread non-communicable disease in the world affecting both adults and children is Dental caries, which causes tooth decay or dental cavities. Dental plaque, which adheres to the tooth surface as a soft gelatinous mass comprises of bacteria embedded in an organic matrix. Streptococcus species are among the first to colonize a tooth surface, thereby initiating the formation of dental plaque, which eventually may cause caries23. S. mutans and S. sobrinus, the two members of Streptococcus species, are the most prevalent cariogenic bacteria found in humans and are considered the principal etiologic agents of dental caries disease24. The identification of anaerobic cariogenic bacteria by the standard traditional culture method is not always sensitive enough and requires special technical skill while isolating fastidious anaerobic bacteria. The main goal of this study was to develop a culture-independent diagnostic test to verify the presence of the microorganisms in dental plaques. Bacterial small subunit 16S rRNA gene is used to detect bacterial pathogens. The Semi-quantitative Real-Time PCR amplification of the 16S rRNA gene using 16S rRNA universal primers confirmed that the purified genomic DNA obtained from the dental plaque samples was indeed of bacterial origin.
Previous studies have revealed that RT-PCR detection is an effective method as compared to other biochemical tests to distinguish S. mutans from other oral Streptococci species25. RT-PCR method is quick, relatively simple, and low concentrations of template DNA is sufficient for the analysis. Previously, a comparative study was conducted to detect the presence of S. mutans in the saliva of children with that of adults and adolescents25. They concluded that the saliva samples harvested from children contained significantly lower S. mutans as compared to saliva samples from adults and adolescents. However, plaque samples collected from children were found to contain S. mutans noticeably more frequently than plaque samples collected from adolescents and adults25.
In the present study, Semi-quantitative Real-time PCR analysis using species specific glucosyltransferase primers was performed in order to identify S. mutans and S. sobrinus species. All the five dental plaque samples were individually analyzed for the presence of the two targeted bacterial species. The purpose of this study was to only confirm the presence of the two predominant cariogenic Streptococcus species using Real-time PCR analysis. Approximately 10 – 20 nano grams of the template bacterial genomic DNA was sufficient in the reaction mixture for detecting the two major Streptococcus species using the cost effective qRT-PCR method. In order to identify the non culturable and other diverse bacterial species from the bacterial genomic DNA isolated from dental plaque samples access to a gene sequencer was essential. To assess the complete composition of the bacterial species present in a dental plaque sample high cost new molecular biology techniques like 16S rRNA gene next generation sequencing (NGS), whole-genome sequencing (WGS) or whole metagenome shotgun sequencing needs to be performed.
Conclusion
Firstly, Genomic DNA was purified from the dental plaque samples using the Biopro spin column and magnetic beads kit. DNA purification using spin columns produced better quantity and quality of DNA than using magnetic beads. The purified Genomic DNA was confirmed whether it is of bacterial origin by Semi-quantitative Real-time PCR detection of the bacterial 16S rRNA gene using the universal 16S rRNA primers. Semi-quantitative real-time PCR was used to detect the presence of both the cariogenic Streptococcus species (S. mutans and S. sobrinus) using the S. mutans gtfB and S. sobrinus gtfI gene specific primers and SYBR Green fluorescent dye for real-time detection. Real-time PCR is a feasible non-invasive diagnostic method for microbial community analysis, helps in the early detection of dental caries disease, and the results can be obtained within 2 hours.
Ackowledgement
The authors wish to thank the Managing Director of Alchem Diagnostics Mr. Marappan Ganesan and the Management of PSGR Krishnammal College for Women for utilizing the lab facilities. We wish to thank Dr. J. Srihari, Professor and HoD, Department of Periodontics, Sri Ramakrishna Dental College and Hospital, Coimbatore for providing us with the dental plaque samples. The biochemical tests for identifying the microorganism were done at the Department of Microbiology, Dr. N.G.P Arts and Science College, Coimbatore. The concentration and purity of genomic DNA was determined using a nanodrop Spectrophotometer at the Plant Genetic Engineering Laboratory, Bharathiar University, Coimbatore. The authors would also like to thank Dr. Suguna Shanmugasundaram, Consultant and Mentor, Department of Biotechnology (PG), PSGR Krishnammal College for Women, Coimbatore for her constant motivation, encouragement and support.
Conflict of interest
There is no conflict of interest
Funding source
There are no funding sources
References
- Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(8):e1002533.
- Dewhirst F. E., Chen T., Izard J., et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002-5017.
- Zhang Y., Fang J., Yang J., Gao X., Dong L., Zheng X., Sun L., Xia B., Zhao N., Ma Z., Wang Y. Streptococcus mutans-associated bacteria in dental plaque of severe early childhood caries. J Oral Microbiol. 2022;14(1):2046309.
- WHO fact sheet: https://www.who.int/news-room/fact-sheets/detail/oral-health
- Marsh P. D. Dental plaque as a microbial biofilm. Caries Res. 2004;38(3):204-211.
- Angarita‐Díaz M. P., Díaz J. A., Tupaz H. A., et al. Presence of Streptococcus dentisani in the dental plaque of children from different Colombian cities. Clin Exp Dent Res. 2019;5(3):184–190.
- Kolenbrander P. E., Palmer R. J., Periasamy S., Jakubovics N.S. Oral multispecies biofilm development and the key role of cell–cell distance. Nat Rev Microbiol. 2010;8(7):471–480.
- Kleinberg I. A mixed-bacteria ecological approach to understanding the role of the oral bacteria in dental caries causation: an alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit Rev Oral Biol Med. 2002;13(2):108-125.
- Seow W. K. Early childhood caries. Pediatr Clin North Am. 2018;65(5):941-954.
- Yano A., Kaneko N., Ida H., Yamaguchi T., Hanada N. Real-time PCR for quantification of Streptococcus mutans. FEMS Microbiol Lett. 2002;217(1):23- 30.
- Lee Y. J., Kim M. H., Kim J. G., Kim J. H. Detection of Streptococcus mutans in human saliva and plaque using selective media, polymerase chain reaction, and monoclonal antibodies. Oral Biol Res. 2019;43(2):121-129.
- Kutsch V. K. Dental caries: an updated medical model of risk assessment. J Prosthet Dent. 2014;111(4):280-285.
- Lemos J. A., Palmer S. R., Zeng L., Wen Z. T., Kajfasz J. K., Freires I. A., Abranches J., Brady L. J. The Biology of Streptococcus mutans. Microbiol Spectr. 2019;7(1):10.
- Matsumoto N. M. Role of Streptococcus mutans surface proteins for biofilm formation. Jpn Dent Sci Rev. 2018;54(1):22-29.
- Abiko Y., Sato T., Mayanagi G., Takahashi N. Profiling of subgingival plaque biofilm microflora from periodontally healthy subjects and from subjects with periodontitis using quantitative real-time PCR. J Periodontal Res. 2010;45(3):389-395.
- Bowen W. H. Dental caries – not just holes in teeth! A perspective. Mol Oral Microbiol. 2016;31(3):228-233.
- Sinsimer D., Leekha S., Park S., Marras S. A., Koreen L., Willey B., et al. Use of a multiplex molecular beacon platform for rapid detection of methicillin and vancomycin resistance in Staphylococcus aureus. J Clin Microbiol. 2005;43(9):4585-91.
- Sato T., Matsuyama J., Kumagai T., Mayanagi G., Yamaura M., Washio J., Takahashi N. Nested PCR for detection of mutans streptococci in dental plaque. Lett Appl Microbiol. 2003;37(1):66-69.
- Montgomerie J. Z., Kalmanson G. M., Guze L. B. The use of the catalase test to detect significant bacteriuria. Am J Med Sci. 1966;251(2):184-187.
- Dahlén G, Hassan H, Blomqvist S, Carlén A. Rapid urease test (RUT) for evaluation of urease activity in oral bacteria in vitro and in supragingival dental plaque ex vivo. BMC Oral Health. 2018;18(1):89.
- Benjaminson M. A., Deguzman B. C., Weil A. J. Voges-Proskauer test: Expeditious techniques for routine use. J Bacteriol. 1964;87(1):234-5.
- Clarke H., Cowan S. T. Biochemical methods for Bacteriology. Gen. Microbiol. 1952;6(1-2):187-197
- Ahl T., Reinholdt J. Detection of immunoglobulin Al protease-induced Fab alpha fragments on dental plaque bacteria. Infection and Immunity. 1991;59(2):563-569.
- Igarashi T., Yamamoto A., Goto N. Rapid identification of mutans Streptococcal species. Microbiology and Immunology. 1996;40(11):867-871.
- Flayyih AS, Hassani HH, and Wali, MH. Detection of biofilm genes (gtf) in Streptococcus mutans isolated from human dental caries. Iraqi Journal of Science 2016;57(1A):104-108.

This work is licensed under a Creative Commons Attribution 4.0 International License.





