Manuscript accepted on : 16-06-2023
Published online on: --
Plagiarism Check: Yes
Reviewed by: Dr. Sabyasachi Banerjee
Second Review by: Dr. Maria Vera Jesus Da Costa
Final Approval by: Dr. Hifzur R. Siddique
Babu M and Geethalakshmi Sundararaman*
and Geethalakshmi Sundararaman*
Department of Biotechnology, Sree Narayana Guru College, Coimbatore, Tamil Nadu, India.
Corresponding Author E-mail:s.geethalakshmi@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3153
ABSTRACT: The current research endeavor involved a meticulous analysis of the expression of MYB genes in the Saccharum officinarum Co86032 cultivar under abiotic stress conditions. The study utilized tailored primers designed to target the ScMYB protein mRNA expressed during abiotic stress like drought, which enabled successful amplification of the ScMYB60 gene in Saccharum officinarum Co86032. To quantify gene expression levels in both leaf and stem tissues, real-time PCR analysis was employed, and the specificity and accuracy of the PCR reaction were ensured through melting temperature analysis. The outcome of this study shows that the specified MYB gene got expressed even on the 18th day of the stress which is a significant advancement in comprehending the role of MYB transcription factors in sugarcane in tolerating drought condition, and its findings may have far-reaching implications in improving sugarcane growth and development and augmenting its resilience to environmental stressors. Future investigations could potentially involve in-depth inquiries into MYB genes in sugarcane and other crops, using a diverse range of methodologies to characterize their function and regulation, with the goal of creating more robust and adaptable crops that can effectively withstand shifting environmental conditions.
KEYWORDS: CT value; FP and RP (forward and reverse primer); PCR; RTPCR; Tm value
Download this article as:| Copy the following to cite this article: Babu M, Sundararaman G. Real-time PCR Analysis of ScMYB Gene Expression in Saccharum officinarum Co86032 under Drought-induced Abiotic Stress Conditions. Biosci Biotech Res Asia 2023;20(3). |
| Copy the following to cite this URL: Babu M, Sundararaman G. Real-time PCR Analysis of ScMYB Gene Expression in Saccharum officinarum Co86032 under Drought-induced Abiotic Stress Conditions. Biosci Biotech Res Asia 2023;20(3). Available from: https://bit.ly/43QWprR |
Introduction
Sugarcane, scientifically known as Saccharum officinarum L., is a tall perennial grass belonging to the family Poaceae. It is one of the most important crops grown worldwide and is cultivated in more than 90 countries, primarily in tropical and subtropical regions. Sugarcane is valued for its high sugar content, which is extracted from the stalks and used in a variety of applications, including the production of sugar, ethanol, and molasses10. Sugarcane, which is a robust crop known for its thick, fibrous stalks rich in sucrose that can reach heights of up to 6 meters, undergoes a process where the stalks are harvested and transported to processing facilities, where they are crushed to extract the sugary juice that is then filtered, purified and ultimately crystallized to yield sugar12. Notwithstanding its significance in sugar production, sugarcane also has a critical role in the production of ethanol, a biofuel that can serve as an alternative to gasoline and is generated by fermenting sugarcane juice or molasses, a byproduct of the sugar refining process, with yeast, thereby producing ethanol that can be employed for several applications, such as a fuel for vehicles, and as a solvent in various industrial processes11. Furthermore, the sugarcane crop is highly esteemed for its efficacy in soil conservation and erosion prevention, owing to its extensive root system that helps in stabilizing soil and preventing erosion, thus making it a preferred crop for planting in areas susceptible to soil erosion. Sugarcane plants are continuously exposed to various stress conditions in their environment, including extreme temperatures, drought, salinity, and more, which trigger intricate changes in gene expression at the transcriptional level 22. This transcriptional reprogramming is driven by a variety of stress-responsive transcription factors, which have been extensively studied in recent years. Despite the progress made in understanding the functions of these transcription factors, much more research is needed to fully comprehend the complex mechanisms underlying plant stress responses. The implications of these findings point towards the conclusion that a thorough comprehension of the mechanisms involved in plant stress responses and tolerance to abiotic stress necessitates a comprehensive analysis of the transcriptional regulation of stress-responsive genes. This highlights the crucial importance of these stress-responsive transcription factors as potential targets for the creation of crops with enhanced ability to withstand abiotic stress 23. Transcription factors (TFs), a diverse class of proteins, serve as critical regulators of gene expression by modulating the rate of transcription of their target genes in response to various environmental and cellular cues. The binding of TFs to specific promoter regions, which are located in the DNA sequence of the target gene’s promoter region, initiates or suppresses the transcription of the gene through the activation or deactivation of upstream signaling cascades. TFs thus act as molecular switches that enable cells to respond and adapt to internal and external stimuli13. Moreover, TFs function as master regulators of transcriptional networks by controlling the expression of multiple genes and coordinating their expression patterns. The discovery of transcription factors that are responsive to stress and their corresponding target genes has elucidated crucial mechanisms that underlie plant responses to stress and their ability to endure abiotic stressors. By means of transcriptional regulation of genes that respond to stress, plants can modulate a wide range of physiological, cellular, and molecular processes, which enable them to cope with and adjust to the changing conditions of their environment17. Through the regulation of genes involved in activities like osmotic adjustment, ROS scavenging, hormone signaling, and pathogen defense, transcription factors, including MYB, NAC, AP2/ERF, and WRKY, can direct intricate and dynamic responses that ultimately determine the extent of stress tolerance and adaptation in plants18. Across the plant kingdom, the MYB family of proteins are present in a wide range of plant species including Arabidopsis, rice, soybean, and maize, and have been found to be involved in many cellular processes, including cell differentiation, cell cycle progression, and responses to both biotic and abiotic stressors19. The MYB protein family, a diverse group of transcription factors, plays critical roles in various biological processes, such as growth, development, and response to environmental stresses24. Extensive research has been conducted on these genes in numerous plant species, revealing their involvement in regulating the expression of genes related to metabolism, signal transduction and secondary metabolism14. Additionally, MYB transcription factors regulate complex molecular networks and signaling pathways, coordinating cellular processes. Given their significance, a detailed understanding of their function, regulation and interactions is crucial. Investigating MYB proteins is expected to offer new insights into plant biology and have implications for improving plant growth, development, and adaptation to stresses. The MYB transcription factor, a crucial protein that modulates gene expression, is a key regulator of plants responses to various environmental stressors such as drought, high salinity, extreme temperatures, and heavy metal toxicity15. In the domain of higher plants, transcription factors with the MYB coding are governed by an extensive super gene family that is categorized into three distinctive subfamilies, namely MYB1R, R2R3MYB, and MYB3R, based on the number of consecutive repeats present within the MYB domain21. Upon activation by stress signals, MYB transcription factors enhance the transcriptional activity of stress-responsive genes, thereby improving a plant’s ability to survive under adverse environmental conditions16. Notwithstanding the vital function of MYB transcription factors in regulating plant growth and their ability to modulate responses to abiotic stressors, the precise role of MYB genes in promoting salinity and drought tolerance in plants is largely unknown. However, when it comes to grasses, sugarcane stands out due to its unique and intricate polyploid genome. Transcription factors (TFs) regulate gene expression in crops, playing a crucial role in genetic improvement. They respond to signal transduction pathways and interact with cis-acting elements to modulate transcription efficiency20. Sugarcane types were studied utilising a high-throughput miRNA deep sequencing approach to investigate the control of gene expression by miRNAs during drought stress in sugarcane25.
The MYB transcription factor family is a broad and varied set of regulatory proteins that are essential for plant growth, development, and stress response. These TFs are distinguished by the MYB domain, a conserved DNA-binding domain that allows them to bind to particular DNA sequences and control the expression of target genes. While substantial study has been undertaken on MYB TFs in numerous plant species, including model plants such as Arabidopsis thaliana, rice, and maize, knowledge on the MYB TF family in sugarcane (Saccharum spp.) is limited. Sugarcane is a worldwide important crop, providing a significant supply of sugar and bioenergy. Understanding the regulatory systems controlling sugarcane development and stress responses is critical for increasing crop production and reducing stress.
Several reasons contribute to the scarcity of knowledge on MYB TFs in sugarcane. First, sugarcane has a complicated polyploid genome, which makes genetic and genomic research difficult. Because of the genome’s extremely repetitive structure and the existence of several homologous genes, it is challenging to precisely identify and characterise certain gene families, including MYB TFs. Furthermore, as compared to model plants, sugarcane has gotten less attention from the research community. The emphasis has been on increasing sugar yield and stress tolerance rather than unravelling the underlying molecular pathways. As a result, sugarcane’s molecular and genetic resources are not as extensive as those accessible for other crops. Despite the scarcity of data, some research have looked at the role of MYB TFs in sugarcane. For example, a few MYB genes in sugarcane cultivars have been found and characterised, and their participation in processes such as sucrose metabolism, disease resistance, and abiotic stress responses has been proposed. These results give preliminary information on the activities of MYB TFs in sugarcane, but further study is needed to properly understand their roles and regulatory networks. Future research efforts should focus on investigating the MYB TF family in sugarcane utilising sophisticated genomic and functional genomic techniques to overcome the information gap. To discover MYB genes, analyse their expression patterns under diverse situations, and explore their direct target genes, high-throughput sequencing technologies such as RNA sequencing and chromatin immunoprecipitation sequencing (ChIP-seq) can be used. Overexpression or knockdown experiments using genetic transformation techniques can also be used to investigate the roles of individual MYB genes. By exploiting current information on MYB TFs in other plant species, comparative genomics methods might give useful insights. Researchers can deduce probable activities of MYB TFs in sugarcane and prioritise prospective genes for future investigation by comparing the MYB gene repertoire and their regulation mechanisms across various plants. Despite the scarcity of available data, research on MYB TFs in sugarcane has the potential to further our understanding of the molecular processes driving sugarcane growth, development, and stress responses. More study and cooperation within the scientific community are required to elucidate the precise activities and regulatory networks of MYB TFs in sugarcane, which will eventually lead to improved crop attributes and increased sugarcane productivity. Despite the extensive research on MYB TFs in various plants, only limited information is currently available about the MYB TF family in sugarcane. This knowledge gap highlights the need for further investigation into the MYB TF family in sugarcane, as it could reveal important insights into the plant’s growth, development and stress response mechanisms20. Techniques such as genetic engineering, which involve overexpression of MYB genes, have demonstrated potential in enhancing drought tolerance in crops. The research conducted on MYB proteins underscores their crucial role in regulating plants adaptive responses to environmental stress, presenting new avenues for crop improvement and increased resilience to environmental changes. In the current investigation, the main objective was to scrutinize the expression of MYB genes in Saccharum officinarum Co86032 through the utilization of primers that were specifically designed to target ScMYB protein mRNAs that gets expressed during stress conditions. These primers demonstrated the ability to facilitate successful PCR amplification, utilizing cDNA that was isolated from both leaf and stem tissue of Saccharum officinarum Co86032. Subsequently, the amplified products were subjected to a highly sensitive and quantitative molecular biology technique, known as real-time PCR analysis. This method enables researchers to amplify and detect DNA or RNA sequences in real-time during the PCR process, thereby allowing for the monitoring of critical parameters such as the cycle threshold (CT) value9. Currently, the assessment of gene expression patterns in plants is commonly accomplished using techniques such as Northern blotting, competitive reverse transcription-polymerase chain reaction (RT-PCR), microarray analysis, or quantitative reverse transcription-polymerase chain reaction (qRT-PCR) 26. But, these techniques have their own liitations28,29,30. Instead, qRT-PCR offers numerous advantages, including heightened sensitivity, enhanced specificity, remarkable accuracy, user-friendly operation and reduced consumption of resources31,32. qRT-PCR helps in detecting mRNA titers or analyzing gene expression in a variety of settings, including distinct species 33,34, developmental stages 35,.36 and response processes to both abiotic and biotic stimuli 26,37. Studies have shown that R2R3-MYB genes showed notable changes in expression during abiotic stress in Gossypium raimondii using RT-PCR techniques during the induction of salt stress39. Similarly, in Malus baccata conserved MYB domain expression was analyzed during cold and drought treatments by quantitative real-time PCR (qPCR) analysis40. Several studies done in Arabidopsis42, Cotton and sunflower38,41 revealed importance of expression analysis by inducing various abiotic stress conditions. These approaches are useful in molecular biology research aiming at understanding the genetic basis of stress tolerance and designing crop enhancement strategies.
Materials and Methods
Plant material collection
Samples of both the leaves and the sett of Saccharum officinarum Co86032 were procured from the ICAR-Sugarcane Breeding Institute located in Coimbatore, Tamil Nadu. The setts were cultivated and maintained in the green house and after 8 weeks they were subjected to drought stress. The leaf and stem samples were collected every week, for five weeks, frozen in liquid nitrogen and stored in -80℃.
Drought stress induction
The sugarcane setts were grown for 8 weeks under controlled conditions. One plant was taken as control which was watered normally and kept at normal environmental condition. Three plants were taken as test, where they were not watered at all during the entire duration of the study. The first day was considered as day 0 and a piece of leaf sample was collected and stored in liquid nitrogen. In the same way, leaf samples were collected on day 3, 6, 9, 12, 15 and 18. After 18th day, the plant died and no sample was available. The morphological characters of the plant such as dryness and yellowness of the leaf, curling of the leaf tip were used to confirm the drought stress induction.
Estimation of physiochemical and biochemical parameters during stress
Biochemical parametric response of Co 86032 to drought stress was studied using various parameters by following standard procedures with slight modifications wherever applicable.
Relative water content (RWC)
The relative water content of the leaf sample was measured according to Yamasaki and Dillenberg43. The RWC of the leaf sample was calculated using the formula RWC (%) = [(FW-DW)/(TW-DW)]*100.
Membrane stability index (MSI)
The membrane stability index of the leaf sample was measured according to standard procedure44. MSI was calculated using the formula EC (%) = [1- (C1/C2)]*100.
Total chlorophyll content
Total chlorophyll content and carotenid content of the control and stress induced sugarcane plants were estimated according to the procedure of Arnon45. The following formulae were used:
Total chlorophyll (mg/l) = 20.2A645 + 8.2A663
Total carotenoid (mg/g of fresh weight) = A470 + (0.114 X A663 – 0.638 X A645)
Proline content estimation
Total proline content of control and stress induced plants were estimated according to Bates et al 46.
Total Soluble Protein estimation
Protein content of the control and stress induced plants was performed according to Bradford method47.
Total Ascorbate content
Total Ascorbate content of the control and stress induced plants was performed according to standard procedure48.
Peroxidase assay
Peroxidase activity in the control and stress induced leaf sample was performed according to Kar and Mishra49.
Lipid peroxidation (MDA content) assay
MDA content of the control and stress induced leaf sample was performed according to Heath and Packer50.
Following the manufacturer’s protocol, the plant RNeasy mini kit (Qiagen)2 was utilized to isolate total RNA from frozen tissues. After the frozen tissues were disrupted and homogenized in 700 μl QIAzol lysis reagent in a clean microcentrifuge tube, the tubes containing the homogenate were placed on the benchtop for 5 minutes before chloroform was added to the tubes and vigorously mixed for 15 seconds, following which the samples were centrifuged for 15 minutes at 12,000 rpm at 4°C, and the upper aqueous phase was then transferred to a new collection tube, where 100% ethanol (525 μl) was added and mixed thoroughly before being pipetted up into an RNeasy Mini spin column that was subsequently centrifuged at 10,000 rpm for 15 seconds at room temperature, and the flow-through was collected and RNeasy Mini spin columns used for total RNA purification. After determining RNA quality and quantity by spectrophotometry and using electrophoresis, the RNA samples were stored at -80 C for further analysis.
Reverse transcription-PCR
After obtaining the isolated total RNA, single-strand cDNA synthesis was performed to analyze the predicted pre-mRNA of MYB. Specifically, 1 microgram of the isolated total RNA was used for the cDNA synthesis, following the instructions provided in the First strand cDNA synthesis kit developed by Clontech3.
Primer Blast – Primer design
This tool utilizes the BLAST algorithm to identify suitable primer sequences that can amplify the target gene with high specificity and efficiency. The designed primers can then be synthesized and used in PCR amplifications to obtain the desired DNA fragments for further analysis4. Designed primers were ordered from Barcode Biosciences and annealing temperatures were examined. In order to ensure optimal annealing specificity, the primer pairs were carefully designed with a length of 24-26 nucleotides, taking into consideration factors such as melting temperature (Tm) and GC content. (Table No: 1)
Enhancement of ScMYB Gene Expression through Gene Coding Sequence Amplification
Once the process of cDNA synthesis was completed, the next step was to amplify the cDNA using the set of primers that had been previously prepared5. However, out of the four pairs of primers that were tested, only the S1F/S1R primer pair produced the desired result. The PCR protocol1 included an initial denaturation step at 94°C for 2 minutes, followed by 30 cycles of denaturation at 94°C for 30 seconds. The annealing step was carried out at 52.2°C, 58°C, or 60°C for 40 seconds, followed by an extension step at 72°C for 1.5 minutes. Finally, a final extension step was carried out at 72°C for 10 minutes. This resulted in an amplicon of approximately 1.5 kilobases in length. (Figure No.1)
Primer Sequences for RT-PCR Analysis
Table 1: Designed Primers sequence FP & RP
|
GenBank ID |
Primer |
FP |
RP |
|
|
ScMYB52 |
HF679466 |
S1 |
ATGGGGCGGTCGCCGTGCTGCGAG |
TCAGTTGGACTCACCCCAGCATTCTG |
|
ScMYB53 |
HF679474 |
S2 |
ATGGGGAGGCACTCCTGCTGTTACAAGC |
TAATGTTAAAAAAGTTAATGGTGAA |
|
ScMYB56 |
HF679484 |
S3 |
ATGGGCCGTAGCCCGTGCTGCGA |
CGGCAGGCTTTTTTCCAGCAGCAG |
|
ScMYB57 |
HF679480 |
S4 |
ATGACCTCAACTCCAAGTGACAAAGCGA |
TTATGTGGATGCCTCGCTTTCAAGAC |
Real-time PCR (qPCR) or (qRT-PCR)
An MX3005P thermocycler (Stratagene/Agilent Technologies) was used to perform quantitative PCR (qPCR) amplification6. The reaction mixture contains the following components and their respective volumes, measured in microliters (μl):
Reverse transcribed ssDNA: 1.0
Real Q-PCR 2X master mix (Amplicon): 10.0
Forward primer ATGGGGCGGTCGCCGTGCTGCGAG : 0.4
Reverse Primer TCAGTTGGACTCACCCCAGCATTCTG : 0.4
RNase-free water: 8.2. The total volume of the reaction mixture is 20.0 μl.
PCR conditions
The RealQ-PCR 2X master mix (Ampliqon)8 was utilized in the experiment, with a volume of 10 ul. The reaction conditions were set as follows: 95°C for 15 minutes, followed by 40 cycles of 95°C for 15 seconds, 60°C for 30 seconds, and 70°C for 45 seconds. To serve as a negative control, the RT reaction mix without reverse transcriptase was also included in the experiment.
Melting Curve Analysis for PCR efficiency7
Following the completion of the PCR cycling, dissociation curves were generated at 95°C to verify the specificity of the PCR amplification. All qPCR reactions were conducted with a total of two technical replicates to ensure the reproducibility and reliability of the results.
Results and Discussion
Physiochemical and biochemical parameters
The direct primary stress involves physiologic damages in membranes, as for instance alterations in the permeability and ion flux. The secondary stress is characterized by the simulation of the osmotic stress (water), what brings, as consequences, dehydration, loss of the leaf turgor, inhibition of the growth, and mineral deficiency.
In conditions of water stress, there is the formation of ROS. These forms include oxygen as hydrogen peroxide (H2O2), superoxide radical (O2–), and the hydroxyl radical (OH–). This reactive oxygen forms are known by oxidize important cellular constituents, such as nucleic acids, lipid and protein membrane (bipolar layer), which could take the cells to death.
Relative water content
Measurements of water content expressed on a tissue fresh or dry mass basis have been mostly replaced by measurements based on the maximum amount of water a tissue can hold. These measurements are referred to as Relative Water Content (RWC). In the present study, RWC decreased gradually and reached threshold value of 44.8% on 18th DOS (Fig. 1).
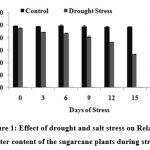 |
Figure 1: Effect of drought and salt stress on Relative water content of the sugarcane plants during stress
|
Membrane stability index (MSI)
The dehydration process during drought is characterized by fundamental changes in water relations, biochemical and physiological processes, membrane structure, and ultrastructure of subcellular organelles. A decrease in membrane stability reflects the extent of lipid peroxidation caused by reactive oxygen species. Premachandra et al51 has reported that cell membrane stability is an indicator of drought tolerance. Fig. 2 indicates the change in MSI during the course of stress induction in both drought and salinity stress conditions of the sugarcane plants. There is a sudden decrease (25%) in the MSI values after 12th DOS in drought stress induced plants. The sudden decrease in MSI indicated that once the plant started expericing severe stress, the membrane integrity decreased and there was a leakage of ions and other cellular components which is reflected in lipid peroxidation and other biochemical characteristics that were estimated..
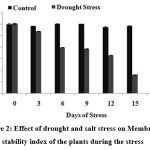 |
Figure 2: Effect of drought and salt stress on Membrane stability index of the plants during the stress
|
Total chlorophyll content
The results showed that there was clear effect of drought on the leaf pigment contents, normal physiology and entire metabolic balance due to stress. It was observed that chlorophyll a/b content increased along the period of time in control plants, while remarkably reduced in stress-induced plants thus proving that drought induced a significant decrease in the contents of pigment fractions (chlorophyll a and b) and consequently the total chlorophyll content (Fig.3). The reduced level of total chlorophyll content (5.13 mg/l for drought stress on the 15th DOS) under stress condition is due to chloroplastid membrane deterioration, leading toward lesser accumulation of chlorophyll and decrease in photosynthetic efficiency.
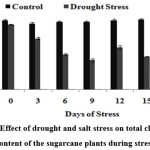 |
Figure 3: Effect of drought and salt stress on total chlorophyll content of the sugarcane plants during stress
|
Total proline content
Proline accumulation in stressed plants is a primary defense response to maintain osmotic pressure in a cell. Proline itself may act not only as an osmoticum but also as a substrate for the TCA cycle during recovery from stress, while the interconversions between proline and its precursors may be involved in the regulation of cellular pH and redox potential.
There was significant increase in proline concentration in stressed plants as compared to its control; however, proline level was noticeably increased on 12th DOS in drought stress treated plants (0.1 µg/mg of fresh weight) (Fig.4).
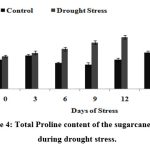 |
Figure 4: Total Proline content of the sugarcane plants during drought stress.
|
Total soluble protein estimation
The main idea underlying studies of stress-induced protein synthesis in plants is that the different sources of stress, their duration, and severity lead to differential expression of genetic information, resulting in changes in gene products, including mRNA and proteins. Such newly synthesized proteins are specific to the particular type of stress and possibly confer enhanced survival value to the plants
The total soluble proteins was increased drastically on 15th day in drought stressed plants (0.5 mg/ml) but with a slight noticeable increase in untreated controls as illustrated in Fig.5, indicating that proteins accumulate during stress.
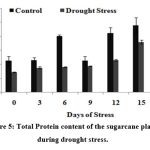 |
Figure 5: Total Protein content of the sugarcane plants during drought stress.
|
Total ascorbate
Production of activated oxygen species is enhanced in plants in response to different environmental stresses such as salinity, drought, waterlogging, temperature extremes, high light intensity, herbicide treatment or mineral nutrient deficiency. Plants containing high concentrations of antioxidants show considerable resistance to the oxidative damage caused by the activated oxygen species.
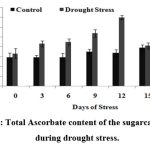 |
Figure 6: Total Ascorbate content of the sugarcane plants during drought stress.
|
Fig.6 shows the ascorbate content of stress induced plants from which it is clear that there was a gradual increase in the ascorbate content of the sugarcane plantlets till 12th DOS (upto 3.5 mg/mg of fresh weight) indicating the defense mechanism adopted by the plant to fight against active oxygen species showing that the ascorbate responses are directly involved in the protection of plant cells against adverse environmental conditions52.
Peroxidase assay
H2O2 plays a dual role in plants: at low concentrations, it acts as a signal molecule involved in acclimatory signaling triggering tolerance to various biotic and abiotic stresses and, at high concentrations, it leads to programmed cell death.
The level of peroxidase showed a gradual increase in drought stressed plants. But the level of peroxidase was higher during drought stress which can be explained as indicating the role of OH– and O2– ions in ROS detoxification
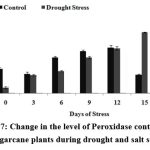 |
Figure 7: Change in the level of Peroxidase content in the sugarcane plants during drought and salt stress.
|
Lipid peroxidation (MDA content) assay
Drought stress results in stomatal closure, which limits CO2 fixation and reduces NADP+ regeneration by the Calvin Cycle. These adverse conditions increase the rate of activated oxygen species (AOS) such as H2O2 (hydrogen peroxide), O2 (superoxide), O2– (singlet oxygen) and OH– (hydroxyl) radicals, by enhanced leakage of electrons to molecular oxygen. These cytotoxic AOS can destroy normal metabolism through oxidative damage to lipids, proteins and nucleic acids.
Generally, it is assumed that salt-induced damage to membranes is negatively correlated with the capacity for increasing activities of enzymes in plants. Fig.8 shows the change in the MDA content of the plants during stress conditions. There was a gradual increase (upto 15 µg/mg of fresh weight) in the case of drought stressed plants.
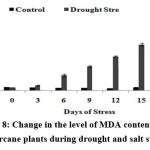 |
Figure 8: Change in the level of MDA content in the sugarcane plants during drought and salt stress.
|
PCR and Real Time PCR Results
Polymerase Chain Reaction (PCR) was conducted using all four pairs of primers S1 – S4. The template utilized in the PCR reaction was complementary DNA (cDNA) extracted from sugarcane. However, only one pair of primers, specifically S1F/S1R, was successful in producing the desired results. The resulting amplicon length was approximately 1.5 kilobases (kb), indicating that the S1F/S1R primer pair was the most effective in amplifying the target region of interest.
 |
Figure 9: Amplicon in lanes 2 and 3.
|
Real time PCR
In order to confirm the amplification or presence of the target gene, real-time polymerase chain reaction (PCR) was conducted. Duplicate samples were analyzed and the results were recorded in the form of amplification curves and melting temperatures. The CT values obtained from the amplification curves for the two duplicate samples were 32 and 31.4, indicating that there was a significant amount of target gene present in the samples. The amplification was evident in the samples collected in the fifth week, which explains that the expression of stress induced gene reaches its maximum during the fifth week of stress induction. Furthermore, no amplification was observed in the negative control, which confirmed that the amplification was specific to the target gene. The melting temperatures for the duplicate samples were 78.99 and 79.29°C, which were consistent with the expected melting temperature for the target gene. Together, these results provide strong evidence for the presence of the target gene in the samples analyzed (Figure No. 10)
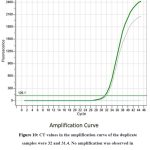 |
Figure 10: CT values in the amplification curve of the duplicate samples were 32 and 31.4. No amplification was observed in the negative control.
|
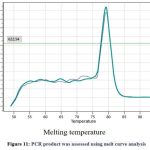 |
Figure 11: PCR product was assessed using melt curve analysis
|
The RT-PCR results indicate that the MYB genes gets expressed during stress conditions in order to protect the plant and to regulate the normal metabolic activities. In Pine tree, the PmMYB4 showed a high level of expression during drought stress and was found to be involved in ABA regulation. Of the 49 sequences selected for analysis, only this MYB gene showed significant expression and was found to be involved in hormonal regulatory pathway42. Similarly, GhMYB73 was found to be showing good expression in Arabidopsis under salt stress condition. Studies done in Malus baccata39 under cold and drought conditions, Gossypium raimondii38 under salt and drought treatment, sunflower under drought conditions, Carica papaya37, rice33 and Brassica34 species, barley27 all have confirmed the importance of MYB genes during stress conditions and their significant contribution in maintaining the hormonal and metabolic balance of the plants during extreme conditions. In all the above-mentioned cases, the life time of the plant varied between 12-20 days after stress induction. Thus, it is clear that MYB class of transcription factors are one of the most important classes of protein family that has a major role in plant growth and development. The current study has also confirmed the fact and can be utilized for overexpression analysis in developing stress resistant crops.
Conclusion
In conclusion, the study has successfully demonstrated the amplification and detection of the ScMYB60 gene in Saccharum officinarum Co86032 through PCR and real-time PCR analysis. The utilization of the S1F/S1R primer pair proved to be effective in amplifying the specific region of interest, resulting in an amplicon length of approximately 1.5 kb. Additionally, the real-time PCR analysis provided further confirmation of the presence of the target gene in the samples, as evidenced by CT values of 32 and 31.4. These values indicate a significant amount of the target nucleic acid present in the initial material, emphasizing the successful detection and quantification of the ScMYB60 gene. The consistent melting temperatures observed in the duplicate samples align with the anticipated melting temperature for the target gene, thereby validating the specificity of the PCR reaction. This specificity assures that the amplified product indeed corresponds to the ScMYB60 gene and is not a result of non-specific amplification.
These findings highlight the practical application of PCR and real-time PCR techniques in the identification and quantification of specific genes in Saccharum officinarum Co86032. As a globally significant crop valued for its high sugar content and ethanol production, as well as its role in soil conservation and erosion prevention, understanding the molecular aspects of this crop is crucial. The MYB family of transcription factors, including ScMYB60, plays a pivotal role in gene regulation and response to environmental stress. The successful detection and amplification of the ScMYB gene using PCR and real-time PCR not only demonstrate the potential of these techniques in studying and improving sugarcane crops but also provide valuable insights into the plant’s stress tolerance. The study reveals that the plant can withstand stress for up to five weeks, after which survival becomes unlikely. The investigation of MYB proteins and their interactions opens new avenues for enhancing crop resilience to environmental changes and promoting agricultural sustainability. By comprehending the molecular mechanisms that govern the expression of vital genes such as ScMYB, we can develop more effective strategies for crop improvement. This knowledge contributes to global food security and sustainability goals.
Furthermore, the isolation and analysis of these genes for overexpression can potentially lead to the enhancement of stress tolerance in industrial crops like sugarcane. Thus, the results of this study hold significant implications for the future of sugarcane genetic engineering and the development of stress-tolerant varieties.
Acknowledgement
I am very thankful to the Department of Biotechnology, Sree Narayana Guru College, K.G. Chavadi, Coimbatore, for providing the necessary facilities to carry out this work.
Conflict of Interest
We declare that we have no conflict of interest.
Funding sources
This research received no particular grant from any funding agency
References
- Lorenz T C. Polymerase chain reaction: Basic protocol plus troubleshooting and optimization strategies. Journal of Visualized Experiments. (63); e3998.
- Vennapusa, A. R., Somayanda, I. M., Doherty, C. J., & Jagadish, S. V. A universal method for high-quality RNA extraction from plant tissues rich in starch, proteins and fiber. Scientific Reports. 10(1); 2020; 1-13.
CrossRef - Matz, M. V., Alieva, N. O., Chenchik, A., & Lukyanov, S. Amplification of cDNA ends using PCR suppression effect and step-out PCR. In J. M. Walker (Ed.), Methods in molecular biology™. 221; 2003; 219-227.
- Ye, J., Coulouris, G., Zaretskaya, I. et al. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 13; 2012; 134.
CrossRef - Matz, M., D. Shagin, E. Bogdanova, O. Britanova, S. Lukyanov, L. Diatchenko, A. Chenchik. Amplification of cDNA ends based on template-switching effect and step-out PCR, Nucleic Acids Research. 27; 6; 1999; 1558–1560.
CrossRef - Ling H, Wu Q, Guo J, Xu L, Que Y. Comprehensive Selection of Reference Genes for Gene Expression Normalization in Sugarcane by Real-Time Quantitative RT-PCR. PLoS ONE 9(5); 2014; e97469.
CrossRef - Pryor, R. J., & Wittwer, C. T. Real-Time Polymerase Chain Reaction and Melting Curve Analysis. In S. K. Kim (Ed.), Methods in molecular biology™. 336; 2006; 63-75.
- Sadasivam, B., & Gajjeraman, P. Characterization of water-deficit stress-responsive R2R3-MYB transcription factor gene, ScMYB50/23 from sugarcane. Indian Journal of Science and Technology. 9(37); 2016;1-8.
CrossRef - Ririe, K. M., Rasmussen, R. P., & Wittwer, C. T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Analytical Biochemistry. 245(2); 1997; 154-160.
CrossRef - Chinnadurai, C. Potential Health Benefits of Sugarcane. In: Mohan, C. (eds) Sugarcane Biotechnology: Challenges and Prospects. Springer; Cham; 2017.
CrossRef - Murali, P., Ram, B., Prathap, D. P., Hari, K., & Venkatasubramanian, V. Sugarcane Based Ethanol Production for Fuel Ethanol Blending Program in India. 2021.
- Singh, A., Lal, U. R., Mukhtar, H. M., Singh, P. S., Shah, G., & Dhawan, R. K. Phytochemical profile of sugarcane and its potential health aspects. Pharmacognosy reviews. 9(17); 2015; 45.
CrossRef - Bhargava, S., Sawant, K. Drought stress adaptation: metabolic adjustment and regulation of gene expression. Plant breeding. 2013;132(1): 21-32.
CrossRef - Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant signaling & behavior. 11(1); 2016; e1117723.
CrossRef - Manna, M., Thakur, T., Chirom, O., Mandlik, R., Deshmukh, R., & Salvi, P. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Plant, Cell & Environment. 44(1); 2020; 1-14.
- Erpen, L., Devi, H.S., Grosser, J.W. et al. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tiss Organ Cult. 132; 2018; 1–25.
CrossRef - Chen, L., Song, Y., Li, S., Zhang, L., Zou, C., & Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 1819(2); 2012; 120-128.
CrossRef - Agnieszka Janiak, Mirosław Kwaśniewski, Iwona Szarejko, Gene expression regulation in roots under drought, Journal of Experimental Botany. 67; 4; 2016; 1003–1014.
CrossRef - Jin, H., & Martin, C. Multifunctionality and diversity within the plant MYB-gene family. Plant molecular biology. 41; 1999; 577-585.
CrossRef - Javed, T., Shabbir, R., Ali, A., Afzal, I., Zaheer, U., & Gao, S. J. Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants. 9(4); 2020; 491.
CrossRef - Liao, Y., Zou, H. F., Wang, H. W., Zhang, W. K., Ma, B., Zhang, J. S., & Chen, S. Y. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell research. 18(10); 2008; 1047-1060.
CrossRef - Ahuja, I., de Vos, R. C., Bones, A. M., & Hall, R. D. Plant molecular stress responses face climate change. Trends in plant science.15(12); 2010; 664-674.
CrossRef - Yoon, Y., Seo, D. H., Shin, H., Kim, H. J., Kim, C. M., & Jang, G. The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants Agronomy. 10(6); 2020; 788.
CrossRef - Yang, X., Zhou, T., Wang, M., Li, T., Wang, G., Fu, F., & Cao, F. Systematic investigation and expression profiles of the GbR2R3-MYB transcription factor family in ginkgo (Ginkgo biloba L.). International Journal of Biological Macromolecules, 172; 2021; 250-262.
CrossRef - Selvi, A., Devi, K., Manimekalai, R., et al. High-throughput miRNA deep sequencing in response to drought stress in sugarcane. 3 Biotech, 11(6); 2021; 312.
CrossRef - Cai J, Li P, Luo X, Chang T, Li J, Zhao Y, et al. Selection of appropriate reference genes for the detection of rhythmic gene expression via quantitative real-time PCR in Tibetan hulless barley. PLoS ONE 13(1); 2018; e0190559.
CrossRef - Hua, W., Zhu, J., Shang, Y., Wang, J., Jia, Q., & Yang, J. Identification of suitable reference genes for barley gene expression under abiotic stresses and hormonal treatments. Plant molecular biology reporter. 33, 2015; 1002-1012.
CrossRef - Fehr, J. E., Trotter, G. W., Oxford, J. T., & Hart, D. A. Comparison of Northern blot hybridization and a reverse transcriptase-polymerase chain reaction technique for measurement of mRNA expression of metalloproteinases and matrix components in articular cartilage and synovial membrane from horses with osteoarthritis. American journal of veterinary research. 61(8); 2000; 900-905.
CrossRef - Pagliarulo, V., George, B., Beil, S. J., Groshen, S., Laird, P. W., Cai, J. & Datar, R. H. Sensitivity and reproducibility of standardized-competitive RT-PCR for transcript quantification and its comparison with real time RT-PCR. Molecular cancer. 3(1); 2004; 1-11.
CrossRef - Choi, W. Y., Kim, J. S., Park, S. J., Ma, C. J., & Lee, H. Y. Microarray-based gene expression profiling to elucidate effectiveness of fermented Codonopsis lanceolata in mice. International journal of molecular sciences. 15(4); 2014; 5907-5915.
CrossRef - Demidenko, N. V., Logacheva, M. D., & Penin, A. A. Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PloS one. 6(5); 2011; e19434.
CrossRef - Derveaux, S., Vandesompele, J., & Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods. 50(4); 2010; 227-230.
CrossRef - Lu, J., Sivamani, E., Azhakanandam, K., Samadder, P., Li, X., & Qu, R. Gene expression enhancement mediated by the 5′ UTR intron of the rice rubi3 gene varied remarkably among tissues in transgenic rice plants. Molecular Genetics and Genomics. 279; 2008; 563-572.
CrossRef - Wiesner, M., Hanschen, F. S., Schreiner, M., Glatt, H., & Zrenner, R. Induced production of 1-methoxy-indol-3-ylmethyl glucosinolate by jasmonic acid and methyl jasmonate in sprouts and leaves of pak choi (Brassica rapa ssp. chinensis). International journal of molecular sciences. 14(7); 2013; 14996-15016.
CrossRef - Koo, S. C., Bracko, O., Park, M. S., Schwab, R., Chun, H. J., Park, K. M., … & Kim, M. C. Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS‐box Gene AGAMOUS‐LIKE6. The Plant Journal. 62(5); 2010; 807-816.
CrossRef - Vaucheret, H., Vazquez, F., Crété, P., & Bartel, D. P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & development. 18(10); 2004; 1187-1197.
CrossRef - Pan, L. J., & Jiang, L. Identification and expression of the WRKY transcription factors of Carica papaya in response to abiotic and biotic stresses. Molecular biology reports. 41; 2014; 1215-1225.
CrossRef - He, Q., Jones, D. C., Li, W., Xie, F., Ma, J., Sun, R., … & Zhang, B. Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Scientific reports. 6(1); 2016; 1-14.
CrossRef - Yao, C., Li, X., Li, Y., Yang, G., Liu, W., Shao, B., … & Han, D. Overexpression of a Malus baccata MYB transcription factor gene MbMYB4 increases cold and drought tolerance in Arabidopsis thaliana. International Journal of Molecular Sciences. 23(3); 2022; 1794.
CrossRef - Zhao, Y., Yang, Z., Ding, Y., Liu, L., Han, X., Zhan, J., … & Ge, X. Over-expression of an R2R3 MYB Gene, GhMYB73, increases tolerance to salt stress in transgenic Arabidopsis. Plant science. 286; 2019; 28-36.
CrossRef - Ahmad, H. M., Rahman, M. U., Ahmar, S., Fiaz, S., Azeem, F., Shaheen, T., … & Mora-Poblete, F. Comparative genomic analysis of MYB transcription factors for cuticular wax biosynthesis and drought stress tolerance in Helianthus annuus L. Saudi Journal of Biological Sciences. 28(10); 2021; 5693-5703.
CrossRef - Lou, X., Yao, S., Chen, P., Wang, D., Agassin, R. H., Hou, Y., … & Ji, K. Transcriptome Identification of R2R3-MYB Gene Family Members in Pinus massoniana and PmMYB4 Response to Drought Stress. Forests. 14(2); 2023; 410.
CrossRef - Yamasaki, S., Dillenburg, L. R. Measurements of leaf relative water content in Araucaria angustifolia. Rev. Bras. Fisiol. Veg. 1999;11(2):69-75.
- .Shanahan, J. F., Edwards, I. B., Quick, J. S., Fenwick, J. R. Membrane thermostability and heat tolerance of spring wheat. Crop Sci. 1990;30(2):247-251.
CrossRef - Arnon, D. I. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant physiol. 1949;24(1):1.
CrossRef - Bates, L. S., Waldren, R. A., Teare, I. D. Rapid determination of free proline for water-stress studies. Plant soil. 1973;39:205-207.
CrossRef - Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analyt. Biochem. 1976;72: 248–254.
CrossRef - Hochberg, M., Melnick, D., Oser, B. Photometric determination of reduced and total ascorbic acid. J Ind Eng Chem. 1943;15(3):182-188.
CrossRef - Kar, M., Mishra, D. Catalase, peroxidase, and polyphenoloxidase activities during rice leaf senescence. Plant physiol. 1976;57(2):315-319.
CrossRef - Heath, R. L., Packer, L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968;125(1):189-198.
CrossRef - Premachandra, G. S., Saneoka, H., Kanaya, M., Ogata, S. Cell membrane stability and leaf surface wax content as affected by increasing water deficits in maize. J. Exp. Bot. 1991;42(2):167-171.
CrossRef - Caverzan, A., Passaia, G., Rosa, S. B., Ribeiro, C. W., Lazzarotto, F., & Margis-Pinheiro, M. (2012). Plant responses to stresses: role of ascorbate peroxidase in the antioxidant protection. Genetics and molecular biology, 35, 1011-1019.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





