How to Cite | Publication History | PlumX Article Matrix
Katyala Srilaxmi and Srinivas Munjam*
and Srinivas Munjam*
Department of Microbiology, Kakatiya University, Warangal, Telangana India.
Corresponding Author E-mail: munjam17@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3146
ABSTRACT: With an overall incidence of over 10% within regular population, Chronic kidney disease is an issue that is becoming more and more important in terms of public health. The enhanced risk of infection, especially those brought on by bacteria that are multi-drug resistant, is one of the main side effects of chronic kidney disease. It is essential to screen and identify multidrug resistant bacteria in chronic kidney disease patients, especially those receiving haemodialysis, in order to prevent the transmission of these pathogens. Hence, to improve outcomes for chronic kidney disease patients, early diagnosis and prompt treatment of drug-resistant bacteria are essential. A total of 2219 samples were screened for antibiotic resistant microbes in hospital samples. 445 samples tested positive (20.05 %) for bacterial growth and 1774 samples tested negative (79.94 %). The rate of multidrug resistance bacterial infections was 17% and 43% higher in CKD patients for estimated glomerular filtration rate between 30 and 59 ml/min/1.73m2 and glomerular filtration rate 30 ml/min/1.73m2 respectively. Five bacterial isolates were found to exhibit multi-antibiotic resistance. The Multiple Antibiotic Resistance (MAR) Index ranged from 0.3 to 0.7 across the isolates. The isolates were identified as Enterobacter bugandensis, Enterococcus faecium, Providencia stuartii, Klebsiella variicola, and Escherichia coli by 16S rRNA gene sequencing and phylogenetic analysis. In conclusion, screening and identification of multidrug resistance bacteria is essential to prevent and control the spread of these pathogens and will be helpful for the effective treatment of the multidrug resistance in chronic kidney disease patients.
KEYWORDS: Antibiotic; Chronic Kidney Disease; Estimated Glomerular Filtration; Multidrug Resistance Bacteria
Download this article as:| Copy the following to cite this article: Srilaxmi L, Munjam S. Screening and Characterization of Multidrug Resistant Bacteria from Chronic Kidney Disease Patients of Warangal. Biosci Biotech Res Asia 2023;20(3). |
| Copy the following to cite this URL: Srilaxmi L, Munjam S. Screening and Characterization of Multidrug Resistant Bacteria from Chronic Kidney Disease Patients of Warangal. Biosci Biotech Res Asia 2023;20(3). Available from: https://bit.ly/3F4g51w |
Introduction
Millions of people globally are impacted by the global health issue known as chronic kidney disease (CKD). During time, there is a cumulative and permanent decrease of kidney function, which causes waste materials and fluid to build up in the body. In the ageing population, the increase in chronic diseases like diabetes and hypertension, as well as other variables including environmental pollution and lifestyle habits, are all contributing to an increase in the prevalence of CKD, which is correlated with substantial mortality and morbidity 1,2.
Recent research has demonstrated that CKD, which affects an estimated 10% of adults worldwide, is a serious global health issue3. By 2040, it is anticipated that CKD, would rank as the 5th leading cause of death globally3. Dialysis with CKD is a multifactorial disease with many risk factors, such as age, sex or race, genetic makeup, environmental exposures, and lifestyle choices. Also, it is a major contributor to coronary artery disease (CAD), heart failure, and stroke. It is also linked to a higher likelihood of other chronic conditions like diabetes, hypertension, and cancer4. Obesity, smoking, a poor diet, inactivity, and exposure to pollutants from the environment are other CKD risk factors4,5.
Moreover, CKD can lead to mental decline, sadness, and a diminished standard of living4. Individuals with CKD frequently have impaired immune systems, making them more prone to bacterial infections. The rise in (MDR) has, regrettably, made it more difficult to treat these infections, which has increased the mortality and morbidity rates6,7.
The MDR bacteria are those that are challenging to eradicate with traditional antibiotic therapy because they are resistant to a minimum of three distinct groups of antimicrobial drugs. CKD patients who have previously compromised immune systems and/or have had kidney transplants are at an increased risk of developing infections, making these germs a serious hazard to them8. According to previous studies, CKD patients had higher rates of MDR bacteria than the general population, with infections of the urinary tract representing the most common infection type9.
A multidisciplinary strategy, including the utilisation of infection control procedures, antimicrobial stewardship programmes, and customised treatment plans, is necessary for the treatment of MDR bacteria in CKD patients. The MDR bacteria can spread among these patients, but the prudent use of antibiotics and the disease management to prevent infections, like hand cleanliness and correct catheter care, can assist10,11. In this study, we aimed to screen and identify multidrug-resistant bacteria from (CKD) patients undergoing dialysis besides exploring the impact of MDR bacteria on CKD patients with urinary tract infections (UTI). The aim of this study was to examine how MDR bacteria affect chronic kidney disease patients and the difficulties in controlling these infections.
Materials and Methods
Sampling and study setting
During the research period, urine samples from 2219 patients with (CDK) who had undergone dialysis were obtained from 7 different dialysis centres in Warangal, Telangana State, India. Demographic details are shown in Figure 1. The samples were obtained between March 2018 and February 2019. Clear-catch mid-stream urine samples were collected from patients, both men and women with ages ranging from 20-65. The samples were taken in 2 ml sterilised screw-cap bottles, kept at 4°C, and then brought right away to the lab for further examination.
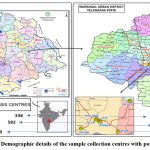 |
Figure 1: Demographic details of the sample collection centres with positive cases.
|
Renal function estimation
This study used electronic health records (EHR) and was an experimental approach. The goal of this study was to ascertain the prevalence of MDR bacteria and the incidence of antibiotic resistance among CKD admitted patients in dialysis centres of Warangal. Serum creatinine concentration and infections that were confirmed by microbial culturing at the time of hospitalization were taken into consideration for being included in the research. Patients receiving kidney replacement therapy who were also terminally sick were not included in the study. The estimated glomerular filtration rate (eGFR) of four major CKD Epidemiology Collaboration (KD-EPI) categories – eGFR 105, 60-104, 30-59, and 30 ml/min/1.73 m2 m2 was taken into consideration and compared for the reason that this range had the lowest infection risk and highest eGFR in an earlier study12.
Isolation of bacteria from urine samples
On different media viz, blood agar, nutrient agar and McConkey agar, bacterial colonies were isolated using spread plate method. The samples collected were diluted by 104-fold using sterilized water and 0.1ml of diluted sample was then spread quantitatively on petri plates having different media. The plates were finally incubated at 37⁰C for 24 for the growth of bacterial colonies.
For this study, standard reference strains viz., E. coli MTCC-443, K. pneumoniae MTCC–4031, and P. aeruginosa MTCC-1688, procured from the MTTC, IMTECH, Chandigarh were used in this present study.
Preparation of the test organisms for antibiogram profiling
Conforming to the guiding principles of the CLSI (Clinical and Laboratory Standards Institute), only 36 morphologically distinct bacterial colonies were evaluated for antibiotic sensitivity pattern by disc diffusion Kirby methodology on MH (Mueller-Hilton) agar13. Fresh bacterial culture in an aliquot of 0.1 ml and 0.5 McFarland was placed on MH agar and left to dry for 5 to 10 minutes. Following that, antibiotic discs were positioned in petri plate and incubated for 24 hours at 37⁰C14. The tested antibiotics like Ampicillin (2mcg), Ceftriaxone (30mcg), Levofloxacin (5mcg), Tazobactam (10mcg), Gentamycin (10mcg), Meropenem (10mcg), Imipenem (10mcg), Co-trimoxazole (25mcg), Tobramycin (30mcg), Norfloxacin (10mcg), Methicillin (10mcg), Azithromycin (15mcg), Ceftazidime (30mcg), Cefoxitin (30mcg), Cefazolin (30mcg), Ciprofloxacin (5mcg), Ticarcillin-Calvunate (15mcg), Tetracycline (30mcg), Fosfomycin (200 mcg) from Himedia, Mumbai, were procured. Following a 24 h incubation, the diameter (mm) of the inhibition was measured using the meter ruler and recorded in accordance with CLSI15. Only one of the isolates is subsequently studied if two or even more isolates collected from the same sample site showed identical results.
Determination of antibiotic resistance and multidrug resistance pattern
The Multi-Drug Resistant Bacteria (MDR) were those bacteria that have demonstrated resistance to more than 12 different antibiotics. The MAR index value for each bacterial isolate was calculated based on the formula.
MAR Index Value = M/n
Where, M is the number of drugs/antibiotics to which the bacterial isolate shows resistance; n is the total number of drugs/antibiotics employed16. Mostly, MAR index greater than 0.2 is an indication that the isolate is MDR17.
Cultural and biochemical characterization of MDR isolates
MDR bacterial isolates obtained from MAR index were further characterized by both cultural and biochemical tests according to the Bergey’s Manual of Systematic Bacteriology18. Cell morphology was microscopically observed using Gram staining. Biochemical characteristics viz., IMViC, oxidase, nitrate reductase, H2S production, sugar fermentation, catalase, amylase and gelatin hydrolysis, etc. were carried out19.
Molecular identification of MDR isolates
DNA isolation and PCR amplification
DNA from 24 h old bacterial isolates was extracted using phenol-chloroform and stored at 16⁰C20. Using a set of forward primer, 27F (50′-AGAGTTTGATCCTGGTCAG-3′) and reverse primer, 1492 R, the 16S rRNA genes were amplified (5′- GGTTACCTTGTTACGACTT-3’) After PCR amplification, DNA was purified by QIAGEN QIAquick PCR Purification kit (Cat.No./ID:28104) and 15 ml of the amplicons were electrophoresed on 1% agarose gel for 1hr at 80 Volts. The DNA bands were then visualized under a UV-transilluminator and the purified product was sent for sequencing21.
16S rRNA gene sequencing and phylogenetical analysis
The sequencing of 16S rRNA gene of the bacterial isolates was performed by Sanger’s method using Applied Biosystems 3730xL analyser by Barcode Biosciences, Bangalore India. Using the BLAST programme, the FASTA sequences were further analysed for taxonomical identification by comparing them to a database of 16S rRNA sequences from NCBI22. With the use of the NJ (Neighborhood Joining) method, a phylogenetic tree was generated for the alignments of the eight closely related matches with the query sequences23.
Results and Discussion
Initially, the urine samples were assessed for the amount of serum creatinine and eGFR and a different pattern was noted in the first positive culture, according to different eGFR categories. Gram positive MDR bacteria increased while Gram negative MDR bacteria decreased from eGFR between 60–104 ml/min/1.73 m2. Enterococcus sp., were the predominant MDR bacteria, followed by Escherichia sp., Klebsiella sp., Enterobacter sp., and Providencia sp. Compared with eGFR between 60 -104 ml/min/1.73 m2, the proportion of E. coli started to rise as the eGFR declined.
The crude MDR ratio increased with decreasing eGFR for E. coli, Klebsiella, Enterobacter sp., Enterococcus sp., but decreased for Providencia sp. These results are in relation with the studies conducted by Evans and his coworkers24. Also, in a study by two different research teams lead by James and Dalrymple, it was found that eGFR ≥ 105 ml/min/1.73 m2 might indicate malnutrition, which would be predisposed to high risk of infection12,25.
The results from various hospitals over the course of a year indicate varying numbers of samples screened and tested positive for antimicrobial resistance (AMR). Hospital M.G.M screened 764 samples, with 151 testing positive for AMR. Similarly, S.V.R screened 582 samples, of which 119 were positive. Max Care screened 338 samples, yielding 96 positive cases. Hospital Jaya had 11 positive results out of 69 samples. Hospital Rohini, Relief, and Vishwas recorded 10, 10, and 48 positive cases out of 99, 51, and 316 screened samples respectively. In total, 445 samples (20.05%) tested positive for AMR, while 1774 samples (79.94%) tested negative, culminating in a cumulative screening of 2219 samples (Table 1, Figure 2).
Table 1: Screening of positive urine samples for Bacteria from CKD Patients undergoing Dialysis
|
Name of Hospital |
M.G.M |
S.V. R |
Max Care |
Jaya |
Rohini |
Relief |
Vishwas |
|
|
March |
No. of Samples |
51 |
44 |
18 |
7 |
6 |
7 |
17 |
|
No. of Samples |
12 |
11 |
7 |
0 |
2 |
1 |
3 |
|
|
April |
No. of Samples |
54 |
39 |
16 |
5 |
7 |
6 |
24 |
|
No. of Samples |
11 |
8 |
11 |
1 |
0 |
1 |
6 |
|
|
May |
No. of Samples |
52 |
34 |
18 |
3 |
13 |
4 |
31 |
|
No. of Samples |
9 |
6 |
7 |
1 |
2 |
1 |
6 |
|
|
June |
No. of Samples |
49 |
31 |
22 |
10 |
9 |
5 |
26 |
|
No. of Samples |
11 |
11 |
9 |
1 |
1 |
1 |
2 |
|
|
July |
No. of Samples |
57 |
38 |
21 |
5 |
8 |
4 |
25 |
|
No. of Samples |
13 |
13 |
6 |
2 |
1 |
1 |
3 |
|
|
August |
No. of Samples |
50 |
46 |
21 |
0 |
9 |
2 |
26 |
|
No. of Samples |
12 |
8 |
5 |
0 |
1 |
2 |
4 |
|
|
September |
No. of Samples |
51 |
42 |
21 |
6 |
6 |
0 |
27 |
|
No. of Samples |
14 |
12 |
4 |
1 |
0 |
0 |
6 |
|
|
October |
No. of Samples |
55 |
44 |
25 |
2 |
7 |
3 |
24 |
|
No. of Samples |
14 |
13 |
8 |
0 |
0 |
2 |
5 |
|
|
November |
No. of Samples |
52 |
41 |
21 |
3 |
0 |
5 |
15 |
|
No. of Samples |
17 |
11 |
11 |
0 |
0 |
1 |
3 |
|
|
December |
No. of Samples |
56 |
40 |
14 |
5 |
6 |
2 |
19 |
|
No. of Samples |
12 |
10 |
11 |
2 |
1 |
0 |
3 |
|
|
January |
No. of Samples |
48 |
38 |
30 |
6 |
10 |
3 |
19 |
|
No. of Samples |
15 |
7 |
9 |
1 |
1 |
0 |
4 |
|
|
February |
No. of Samples |
49 |
37 |
24 |
7 |
9 |
1 |
17 |
|
No. of Samples |
11 |
9 |
8 |
2 |
1 |
0 |
3 |
|
|
Total samples Screened(N) |
764 |
582 |
338 |
69 |
99 |
51 |
316 |
|
|
No. of Positive Samples(n) |
151 |
119 |
96 |
11 |
10 |
10 |
48 |
|
|
Cumulative Number of Samples (N) |
2219 |
|||||||
|
Total No. of Samples Tested Positive for AMR |
445 (20.05 %) |
|||||||
|
Total No. of Samples Tested negative for AMR |
1774 (79.94 %) |
|||||||
 |
Figure 2: Screening of positive samples from CKD patients undergoing dialysis.
|
36 colonies with unique morphology were picked up and tested for antibiotic sensitivity or resistance. The Multi Antibiotic Resistance (MAR) index is an indication of the bacterial isolates showing resistance to different antibiotics. If the MAR index value is more than 0.2 (>0.2), the bacteria is considered as multi-drug-resistant bacteria. From the MAR indices, it was found that 5 out of 36 isolates exhibited resistance to multiple antibiotics (Figure 3, Table 2).
 |
Figure 3: Disc diffusion antibiotic testing of isolates
|
Table 2: Antibiogram and MAR Index of MDR bacterial Isolates
| Antibiotic Tested |
Bacterial isolate |
|||||
| Class of Antibiotics |
Antibiotic (mcg) |
CKD_07_1 |
CKD_19 |
CKD_21 |
CKD_27_1 |
CKD_31_1 |
|
Ampicillin |
Ampicillin-2 |
R |
S |
S |
R |
R |
|
Macrolide |
Azithromycin-10 |
S |
R |
R |
R |
R |
|
Cephalosporins |
Ceftriaxone-30 |
R |
R |
S |
R |
R |
|
Cephalexin-20 |
S |
R |
R |
S |
S |
|
|
Ceftazidime-30 |
R |
R |
R |
R |
R |
|
|
Cefoxitin-30 |
R |
R |
S |
R |
R |
|
|
Cefazolin-30 |
R |
S |
R |
R |
R |
|
|
Quinoline |
Norfloxacin-10 |
S |
R |
R |
R |
S |
|
Fluoroquinolone |
Levofloxacin-5 |
R |
R |
R |
R |
R |
|
Ciprofloxacin-5 |
R |
R |
S |
S |
R |
|
|
Beta-lactam |
Tazobactam-10 |
R |
R |
R |
S |
S |
|
Ticarcillin-Calvunate-15/10 |
R |
S |
S |
R |
R |
|
|
Methicillin-10 |
R |
S |
S |
R |
S |
|
|
Aminoglycoside |
Gentamycin-10 |
R |
S |
R |
S |
R |
|
Tobramycin-30 |
R |
R |
R |
S |
R |
|
|
Carbapenem |
Meropenem-10 |
S |
S |
S |
R |
R |
|
Imipenem-10 |
S |
S |
R |
S |
R |
|
|
Tetracycline |
Tetracycline-30 |
S |
R |
S |
R |
S |
|
Sulphonamide |
Co-trimoxazole-25 |
R |
R |
R |
R |
R |
|
Fosfomycin |
Fosfomycin-200 |
S |
R |
R |
S |
S |
|
|
MAR Index |
0.7 |
0.35 |
0.4 |
0.35 |
0.3 |
S = Sensitive; R = Resistant
Isolates viz., CKD_7_1, CKD_19, CKD_21, CKD_27_1 and CKD_31 was found to be resistant to most of the antibiotics. Almost all isolated showed resistance towards Ceftazidime and Levofloxacin. Highest MAR index (0.7) was shown by CKD_07_1 and the least by CKD_31_1 (0.3). CKD_07_1 showed resistance to all the antibiotics except Azithromycin, Norfloxacin, Meropenem, Imipenem, Tetracycline, Cephalexin and Fosfomycin; CKD_19 was found to be sensitive to the Ampicillin, cefazolin, Ticarcillin, Methicillin Gentamycin, Meropenem and imipenem. From the results tabulated in table 2, it is evident that patients with CKD are possibly more likely to have a resistant strain.
In a study conducted by Moges and his co-workers, multidrug resistance was also prevalent in Gram-negative bacteria like E. coli, Enterobacter sp. Citrobacter sp. and Klebsiella sp. and Gram-positive bacteria like Staphylococcus sp. which is in accordance with our study26.
Research efforts have extensively delved into the antibiotic sensitivity profiles of Enterococcus faecalis. Numerous investigations have explored the susceptibility of Enterococcus faecalis to various antibiotics. In a recent study conducted by Khalil et al. (2022), the findings highlighted that a majority of E. faecalis strains exhibited resistance to tetracycline, erythromycin, levofloxacin, and quinupristin-dalfopristin27. In parallel, Samani et al. (2021) conducted another study focused on the prevalence of virulence genes and the antibiotic resistance pattern of Enterococcus faecalis isolated from urinary tract infections in Shahrekord, Iran and found that it was resistant to Norfloxacin, Vancomycin, tetracycline, ciprofloxacin and erythromycin28. This is in complete contradiction to the isolate (CKD_7_01) in our study which was sensitive to Norfloxacin and tetracycline.
Providencia stuartii, in a study conducted by Stock and Wiedemann (1998), showed resistance to polymyxin B, colistin and nitrofurantoin, some aminoglycosides and Penicillins, older cephalosporins, tetracyclines, gentamicin, tobramycin and chloramphenicol29. The results were of our isolate (CKD_19) in accordance to the literature.
CKD_21 also showed resistance to many antibiotics similar to a study conducted by Rodriguez et al. (2019), researchers analyzed the antibiotic susceptibility of Klebsiella variicola isolates. They found variable resistance rates to commonly used antibiotics, including ampicillin, amoxicillin/clavulanate, ciprofloxacin, and gentamicin30. Another study by Feng et al. (2020) investigated the genetic basis of antibiotic resistance in Klebsiella variicola. The research highlighted the presence of various resistance genes, including those conferring resistance to beta-lactams, aminoglycosides, and quinolones31.
In a study by Urbaniak et al (2018), it was found that E. bugandensis displayed resistance to cefazolin, cefoxitin, erythromycin, oxacillin, penicillin, and rifampin. Additionally, concerning ciprofloxacin and gentamicin, the strains exhibited either resistance or intermediate resistance. Notably, with tobramycin, a combination of resistant, intermediate resistant, and susceptible strains was observed which was contradictory to our isolate (CKD_27_01) being sensitive to tobramycin32.
In a study by McGregor et al. (2013), which investigated antibiotic resistance patterns of Escherichia coli urinary isolates from outpatients, it was found that E. coli demonstrated susceptibility to ampicillin, amoxicillin-clavulanate, ciprofloxacin, and nitrofurantoin33. The results are in contract with the finding of McGregor.
The MDR bacterial isolates were further analysed by morphological, biochemical and molecular analysis. Biochemical and cultural characteristics of all isolates CKD_7_1, CKD_19, CKD_21, CKD_27_1 and CKD_31_1 was studied and Table 3 specifies the structural and biochemical characteristics of the isolated strains.
Table 3: Morphological and biochemical characterization of MDR bacterial isolates
|
Morphological and biochemical test |
Name of the MDR bacterial isolate |
||||
|
|
CKD_07_1 |
CKD_19 |
CKD_21 |
CKD_27_1 |
CKD_31_1 |
|
Gram stain |
+ |
– |
– |
– |
– |
|
Motility |
– |
+ |
– |
+ |
+ |
|
Catalase production |
– |
+ |
+ |
+ |
+ |
|
Gelatin hydrolysis |
– |
– |
– |
– |
– |
|
Voges Proskauer |
+ |
– |
+ |
+ |
– |
|
Lipase |
– |
– |
– |
– |
– |
|
Methyl red |
– |
+ |
– |
– |
+ |
|
Oxidase |
+ |
– |
– |
– |
– |
|
Indole production |
+ |
+ |
– |
– |
+ |
|
Citrate |
– |
+ |
+ |
+ |
– |
|
Urea hydrolysis |
– |
+ |
+ |
– |
– |
|
Nitrate reduction |
– |
+ |
+ |
+ |
+ |
|
D-Glucose (acid/gas) |
-/+ |
+/- |
-/+ |
+/+ |
+/+ |
|
Lactose |
+ |
– |
+ |
– |
+ |
|
Sucrose |
+/- |
+/- |
+ |
+ |
+/- |
|
L-Arabinose |
+ |
– |
+ |
+ |
+ |
|
Glycerol |
+ |
– |
+ |
+ |
– |
|
D-Glucoside |
– |
– |
+/- |
+ |
– |
|
Raffinose |
– |
– |
+ |
+ |
+/- |
|
D-Sorbitol |
– |
– |
+ |
+ |
+ |
|
D-Mannitol |
+ |
– |
+ |
+ |
+ |
|
Maltose |
+ |
– |
+ |
+ |
+ |
|
D-Adonitol |
– |
– |
+ |
– |
– |
|
Cellobiose |
+ |
– |
+ |
+ |
– |
The DNA from all the isolated strains was isolated, amplified and the PCR amplicons were analysed for molecular identification by 16S rRNA sequencing employing Sanger’s method. Nucleotide BLAST studies revealed that the 16S rRNA sequences of the isolates, CKD_07_1 (919 bp), CKD_19 (711 bp), CKD_21 (705 bp), CKD_27_1 (875 bp), CKD_31_1 (897 bp) was found to be Enterococcus faecium, Providencia stuartii, Klebsiella variicola, Enterobacter bugandensis and Escherichia coli respectively when checked with nearest homology. The partial sequences of 16s rRNA gene for the MDR isolates were submitted to the GenBank, NCBI and can be accessed under the given accession numbers (Table 4). The phylogenetic trees were constructed for few MDR bacterial isolates (Figure 4).
Table 4: MDR bacterial isolates and their accession numbers
|
Name of the Isolate |
Accession Number |
|
CKD_07_1 |
OQ719832 |
|
CKD_19 |
OQ719835 |
|
CKD_21 |
OQ719895 |
|
CKD_27_1 |
OQ719944 |
|
CKD_31_1 |
OQ719882 |
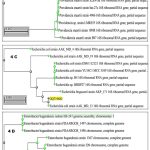 |
Figure 4: Phylogenetic tree of (4.A) CKD_7_1 (4.B) CKD_19 (4.C) CKD_31_1 (4.D) CKD_27_1 (4.E) CKD_21
|
CKD appears to be a health risk for bacterial infection brought on by pathogens that are resistant to antibiotics, but the majority of published studies are concentrated on the risk of MDR bacterial infections in patients with end stage kidney disease (ESKD) who are receiving dialysis, and there are few studies that examine the association between early stages of CKD and antibiotic susceptibility34.
Conclusions
The prevalence of antibiotic-resistant strains, including Enterococcus faecium, Providencia stuartii, Klebsiella variicola, Enterobacter bugandensis, and Escherichia coli, among CKD patients underscores the urgency of addressing antimicrobial resistance. On the other hand, Cephalosporins and the beta lactam class of antibiotics are frequently administered in case of any bacterial infections, which could be the reason for development of resistance in most of the bacteria. In order to prevent the overuse of antibiotics and shorten the treatment times, it is vital to develop efficient guidelines for the judicious usage of antibiotics for CKD patients. Research into plant extracts’ efficacy against MDR bacteria is on-going, where they can also be used to combat multidrug resistant bacteria. By leveraging in silico methods and data analysis, we can identify new drug targets and develop more effective antibiotics to combat MDR infections in CKD patients with UTIs also. Future challenges in this field include the need to improve the accuracy and reliability of in silico analysis, as well as to validate the findings of in silico studies with experimental data that can account for the complex and dynamic nature of multidrug resistance in CKD patients, including the impact of co-morbidities and drug interactions.
Acknowledgment
The authors are grateful to the department of Microbiology, Kakatiya University and thankful to the Head, Department of Microbiology for their facilities and constant support.
Conflict of Interests
The authors declare that there are no conflicts of interest.
References
- Bello, A. K., Nwankwo, E., El Nahas, A. M. Prevention of chronic kidney disease: a global challenge. Kidney International. 2005; 68, pp.S11-S17.
CrossRef - Lv, J. C., & Zhang, L. X. Prevalence and disease burden of chronic kidney disease. Renal fibrosis: mechanisms and therapies. 2019; pp.3-15.
CrossRef - Bikbov, B., Purcell, C. A., Levey, A. S., Smith, M., Abdoli, A., Abebe, M., Owolabi, M. O. Global, regional, and national burden of chronic kidney disease, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. The lancet. 2020; 395(10225), pp.709-733.
- Chu, C. D., McCulloch, C. E., Banerjee, T., Pavkov, M. E., Burrows, N. R., Gillespie, B. W., Waller, L. CKD awareness among US adults by future risk of kidney failure. American Journal of Kidney Diseases. 2020; 76(2), pp.174-183.
CrossRef - Saliba, R., Zahar, J. R., Dabar, G., Riachy, M., Karam-Sarkis, D., Husni, R. Limiting the Spread of Multidrug-Resistant Bacteria in Low-to-Middle-Income Countries: One Size Does Not Fit All. Pathogens. 2023; 12(1), pp.144.
CrossRef - Morehead, M. S., Scarbrough, C. Emergence of global antibiotic resistance. Primary care: clinics in office practice. 2018; 45(3), pp. 467-484.
CrossRef - World Health Organization. Antimicrobial resistance: global report on surveillance. WHO Press. 2019.
- Molnar MZ, Alhourani HM, Wall BM, Lu JL, Streja E, Kalantar-Zadeh K, Kovesdy CP. Association of hepatitis C viral infection with incidence and progression of chronic kidney disease in a large cohort of US veterans. . 2015 May;61(5): pp.1495-502.
CrossRef - McCullough, P. A., Uhlig, K., Neylan, J. F., Pergola, P. E., Fishbane, S. Usefulness of oral ferric citrate in patients with iron-deficiency anemia and chronic kidney disease with or without heart failure. The American journal of cardiology. 2018; 122(4), pp.683-688.
CrossRef - Moradi, H., Vaziri, N. D. Lipid disorders associated with chronic kidney disease and nephrotic syndrome. Endocrine Disorders in Kidney Disease: Diagnosis and Treatment. 2019; pp.153-169.
CrossRef - Algammal, A., Hetta, H. F., Mabrok, M., Behzadi, P. Emerging multidrug-resistant bacterial pathogens “superbugs”: A rising public health threat. Frontiers in Microbiology, 2023; pp.14.
CrossRef - Dalrymple, L. S., Mu, Y., Romano, P. S., Nguyen, D. V., Chertow, G. M., Delgado, C., Johansen, K. L. Outcomes of infection-related hospitalization in Medicare beneficiaries receiving in-center hemodialysis. American Journal of Kidney Diseases.2015; 65(5), pp.754-762.
CrossRef - Bauer, A. W., Kirby, W. M., Sherris, J. C., Turck, M. Antibiotic susceptibility testing by a standardized single disk method. American journal of clinical pathology. 1966; 45(4), pp.493–496.
CrossRef - Fekete, T., Tumah, H., Woodwell, J., Truant, A., Satischandran, V., Axelrod, P., Kreter, B. A comparison of serial plate agar dilution, bauer-kirby disk diffusion, and the vitek automicrobic system for the determination of susceptibilities of Klebsiella spp., Enterobacter spp., and Pseudomonas aeruginosa to ten antimicrobial agents. Diagnostic microbiology and infectious disease. 1994; 18(4), pp.251-258.
CrossRef - Hsueh, P. R., Ko, W. C., Wu, J. J., Lu, J. J., Wang, F. D., Wu, H. Y., Teng, L. J. Consensus statement on the adherence to Clinical and Laboratory Standards Institute (CLSI) Antimicrobial Susceptibility Testing Guidelines (CLSI-2010 and CLSI-2010-update) for Enterobacteriaceae in clinical microbiology laboratories in Taiwan. Journal of Microbiology, Immunology and Infection. 2010; 43(5), pp.452-455.
CrossRef - Krumperman, P. H. Multiple antibiotic resistances indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Applied and environmental microbiology. 1983; 46(1), pp.165-170.
CrossRef - Adefisoye MA, Okoh AI (2017). Ecological and public health implications of the discharge of multidrug-resistant bacteria and physicochemical contaminants from treated wastewater effluents in the Eastern Cape, South Africa. Water. 2017; 9(8): pp.562.
CrossRef - Sharpe, M. Elisabeth, ed. Bergey’s manual of systematic bacteriology. Wiliams and Wilkins, 1986.
- Cappuccino, J. G. S. Microbiology: a laboratory manual/James G., Cappuccino and Natalie Sherman. 1999; (No. 576 C3.).
- Sambrook, J., Fritsch, E. F., Maniatis, T. Molecular Cloning. A Laboratory Manual, 2nd edn.(Cold Spring Harbor Laboratory Press: New York, USA.) 1989.
- Satyanarayana, S. D., Krishna, M. S. R., Kumar, P. P. Optimization of highyielding protocol for DNA extraction from the forest rhizosphere microbes. 3 Biotech. 2017; 7, pp.1-9.
CrossRef - Whitford M.F. Forster R.J. Beard C.E. Gong J Teather R.M. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe. 1998; 4, pp.153–163.
CrossRef - Saitou, N., Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987; 4(4), pp.406-425.
- Evans, R., Caskey, F., Fluck, R., Crowley, L., Davies, J., Nsonwu, O., & Farrington, K. UK Renal Registry 18th annual report: chapter 12 epidemiology of reported infections amongst patients receiving dialysis for established renal failure in England 2013 to 2014: a joint report from Public Health England and the UK Renal Registry. Nephron Clinical Practice. (2016).
CrossRef - James, M. T., Quan, H., Tonelli, M., Manns, B. J., Faris, P., Laupland, K. B., Alberta Kidney Disease Network. CKD and risk of hospitalization and death with pneumonia. American Journal of Kidney Diseases. 2009; 54(1), pp.24-32.
CrossRef - Moges, F., Endris, M., Belyhun, Y., Worku, W. Isolation and characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital environments, Northwest Ethiopia. BMC research notes. 2014; 7, pp.1-6.
CrossRef - Khalil, M. A., Alorabi, J. A., Al-Otaibi, L. M., Ali, S. S., Elsilk, S. E. Antibiotic resistance and biofilm formation in Enterococcus spp. isolated from urinary tract infections. Pathogens. 2022; 12(1), 34.
CrossRef - Samani, R. J., Tajbakhsh, E., Momtaz, H., Samani, M. K. Prevalence of virulence genes and antibiotic resistance pattern in Enterococcus faecalis isolated from urinary tract infection in Shahrekord, Iran. Reports of Biochemistry & Molecular Biology. 2021; 10(1), 50.
CrossRef - Rodríguez-Medina, N., Barrios-Camacho, H., Duran-Bedolla, J., Garza-Ramos, U. Klebsiella variicola: an emerging pathogen in humans. Emerging microbes & infections. 2019; 8(1), 973-988.
CrossRef - Stock, I., and Wiedemann, B. Natural antibiotic susceptibility of Providencia stuartii, P. rettgeri, P. alcalifaciens and P. rustigianii strains. Journal of medical microbiology 1998; 47(7), 629-642.
CrossRef - Li, Shuangshuang, Xudong Feng, Min Li, and Zhen Shen. “In vivo adaptive antimicrobial resistance in Klebsiella pneumoniae during antibiotic therapy.” Frontiers in Microbiology. 2023; 14, 1159912.
CrossRef - Urbaniak, C., Sielaff, A. C., Frey, K. G., Allen, J. E., Singh, N., Jaing, C., … Venkateswaran, K. Detection of antimicrobial resistance genes associated with the International Space Station environmental surfaces. Scientific reports. 2018; 8(1), 814.
CrossRef - McGregor, J. C., Elman, M. R., Bearden, D. T., Smith, D. H. Sex-and age-specific trends in antibiotic resistance patterns of Escherichia coli urinary isolates from outpatients. BMC family practice. 2013; 14(1), 1-5.
CrossRef - Vacaroiu, I. A., Cuiban, E., Geavlete, B. F., Gheorghita, V., David, C., Ene, C. V., Radulescu, D. Chronic Kidney Disease-An Underestimated Risk Factor for Antimicrobial Resistance in Patients with Urinary Tract Infections. Biomedicine. 2022; 10(10), pp.2368.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





