How to Cite | Publication History | PlumX Article Matrix
A Synopsis of the Causes, Hypotheses, Progression, and Genes Associated with Breast Cancer
Department of Biological Sciences, University Collage of Haqel, University of Tabuk, Tabuk, Saudi Arabia.
Corresponding Author Email:halatwi@ut.edu.sa
DOI : http://dx.doi.org/10.13005/bbra/3162
ABSTRACT: Cancers are widespread among women, and breast cancer is considered one of the deadliest cancers among women. Due to early detection of breast cancer and appropriate treatment, the recovery rate among women increases, reaching 80% in developed countries. There are various genes linked to breast cancer as well as cancer stem cells. Genetic and epigenetic changes that result in abnormal gene function are involved in breast cancer. The genes associated with breast and ovarian cancer are BRCA1 and BRCA2. They are tumour suppressor genes because they play a role in DNA repair procedures. Furthermore, different malignancies exhibit dysregulation of the MYC oncogene family, which is typically associated with a bad prognosis for tumours. HER receptors are found in many tissues of epithelial, mesenchymal, and neuronal origin, where they control a variety of vital processes such as cell migration, programmed cell death (apoptosis), cell differentiation, and abnormal cell growth. Our understanding of the mechanisms that give birth to breast cancer has been significantly impacted by recent advances in genomics research. This understanding has in turn prompted the development of novel diagnostic and treatment techniques. This review will focus on the concept of breast cancer and related genes.
KEYWORDS: Breast cancer; BRCA1; BRCA2; c-MYC; HER2
Download this article as:| Copy the following to cite this article: Alatawi H. A. A Synopsis of the Causes, Hypotheses, Progression, and Genes Associated with Breast Cancer. Biotech Res Asia 2023;20(4). |
| Copy the following to cite this URL: Alatawi H. A. A Synopsis of the Causes, Hypotheses, Progression, and Genes Associated with Breast Cancer. Biotech Res Asia 2023;20(4). Available from: https://bit.ly/3RpWjDn |
Introduction
An summary of female breast cancer statistics in the US, including information on incidence, mortality, survival, and screening, is given by the American Cancer Society. In the United States, women are predicted to experience 40,290 breast cancer-related fatalities and 231,840 new instances of invasive breast cancer in 2015. From 2008 to 2012, the incidence rates of breast cancer were steady for non-Hispanic white (white), Hispanic, and American Indian/Alaska Native women, but increased for black and Asian/Pacific Islander women. Despite traditionally having greater incidence rates than black women, white women’s rates converged in 2012. Remarkably, from 2008 to 2012, incidence rates in seven states, mostly in the South, were considerably higher for black women than for white women. There were 249,000 fewer breast cancer deaths in the US between 1989 and 2012 as a result of a 36% decline in breast cancer death rates. With the exception of American Indians and Alaska Natives, all racial and ethnic groupings saw a decline in death rates. However, the mortality gap between white and black women has remained persistent across the country, with black women’s death rates being 42% higher in 2012 than those of white women1.
The difficulty in treating and controlling malignant breast cancer lies in the fact that it extends and spreads to distant organs such as the bones, liver, lungs, and brain. Early diagnosis of breast cancer effectively contributes to avoiding the complications of the disease and reducing the high mortality rate among women . As a result of this early diagnosis, the cure rate increases to 80%, as in North America 2.
One of the most common methods for diagnosing breast cancer is mammography. This method has proven its ability to predict the formation of a mammary tumor, thus speeding up the diagnosis of the disease and reducing the mortality rate. As a result of the advancement of techniques used in diagnosing breast cancer over the past ten years, new screening techniques have been used, such as magnetic resonance imaging (MRI). It is considered more accurate compared to mammography3. The possibility of an increased risk of breast cancer is linked to a group of risk factors, including gender, family history, aging, estrogen, genetic mutations, and lifestyle 4.
Due to the congenital and structural differences between female and male, the incidence of breast cancer is 100 times higher in female than in males. Annually, the incidence of breast cancer in America has increased. Nevertheless, the prevalence of early screenings and cutting-edge medical treatments helps to lower the mortality rate. Currently, in light of medical and biological progress, medications have been manufactured based on individuals genome sequencing technologies, which aim primarily to treat breast cancer. This article addresses the functions of genetic machinery in the onset and recurrence of breast cancer.
What is breast cancer?
Breast cancer is a malignant tumour that originates from breast cells. A malignant tumour is a mass of cancer cells that can infect nearby tissues or metastasis (It extends and affects distant organs in the human body). The breast cancer occurs almost entirely in female, however male can get it, too, but in rare cases. Trillions of live cells comprise the human body. In a healthy body, cells develop, divide, and decompose in a predictable manner. Normal cells divide more quickly in a person’s early years to facilitate growth. After reaching adulthood, the majority of cells only divide to replace damaged or dead cells or to heal wounds. When cells in a particular area of the body begin to multiply uncontrollably, cancer is the result. Despite the fact that there are numerous different types of cancer, all these types always begin as a result out-control growth of aberrant cells5.
DNA damage leads to the development of cancerous cells in cells. Each cell contains DNA, which controls all activity in the cell. Damage to DNA in a normal cell results in one of two outcomes: either the cell repair the damage or dies. Nonetheless, cancer cells do not repair their damaged DNA, and the damaged DNA cells do not die as expected. Instead, these cells continue to produce additional cells that the body does not require. The damaged DNA present in the original cell will be present in all of these subsequent cells. Although DNA damage can be passed down through the generations, most DNA damage is triggered on by environmental factors or mistakes that occur during normal cell reproduction (Fig. 1) 5.
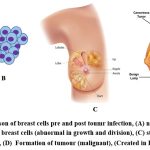 |
Figure 1: Comparison of breast cells pre and post toumr infection, (A) normal breast cells in women, (B) cancer breast cells (abnormal in growth and division) |
Origins of breast cancer
The first step in the formation of a breast tumor is the excessive proliferation of ducts (hyperproliferation), then this growth is stimulated by a group of carcinogenic stimuli, and ultimately this excessive growth turns into either benign or malignant tumors. Breast cancer formation and development are greatly influenced by the tumor microenvironment, which is represented by stromal and macrophage influences. Studies conducted on the mammary glands of mice have shown that tumors develop significantly when the stroma is exposed to carcinogens compared to when it is exposed to the extracellular matrix or epithelium6. Macrophages have the ability to create a mutagenic inflammatory microenvironment that encourages angiogenesis and makes it possible for cancer cells to resist immune rejection7. Different forms of DNA methylation are used to determine differences between normal and tumor-associated microenvironments. Subsequently, it has been proven that epigenetic alteration in the tumour microenvironment can encourage carcinogenesis8.
Recently, secondary malignant cancer cells known as cancer stem cells (CSCs) have been discovered. These cells have an effective role in the beginning of the tumor, its spread to other organs, and the recurrence of the tumor. CSCs originate either from stem cells or ancestor cells within healthy tissues. These cells have the ability to regenerate and are resistant to conventional treatments such as chemotherapy and radiotherapy9. Breast cancer stem cells (bCSCs) were discovered by Ai Hajj. His experiment was based on injecting mice with weak immunity with cancer stem cells (100 cells), and he noticed that the mice’s cells had the ability to form new tumors10. The main source of bCSCs formation is likely to be luminal epithelial stem cells rather than basal stem cells9. Nevertheless, further research is necessary to understand bCSCs and design fresh methods for their complete suppression.
Theories of breast cancer
There are two suppositional theories for the beginning and expansion of breast cancer, the cancer stem cell theory and the stochastic theory11.
The cancer stem cell hypothesis suggests that:
All tumour secondary types are produced by the same stem cells or transit-amplifying cells (progenitor cells). Genetic and epigenetic mutation in stem cells and progenitor cells lead to the emergence of multiple characteristics of tumors.
The stochastic hypothesis suggests that:
Whether the single cell are stem cells, progenitor cells, or differentiated cells, each tumour subtype begins as one of these cell types.
Despite the fact that both hypotheses are well-supported by data, neither of them clearly explain the causes of the occurrence and proliferate of breast cancer.
The breast cancer-related genes
Several genes have been implicated in the onset and development of breast cancer. Oncogene and anti-oncogene have important roles in the initiation and development of tumors. Subsequently, any mutations or abnormal amplification of these genes lead to occurrence of breast cancer (Fig. 2)12.
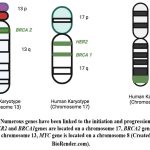 |
Figure 2: Numerous genes have been linked to the initiation and progression of breast cancer. HER2 and BRCA1genes are located on a chromosome 17, BRCA2 gene is located on a chromosome 13, MYC gene is located on a chromosome 8 (Created in BioRender.com). |
Anti-oncogenes: BRCA1 and BRCA2
Two well-known anti-oncogenes for the risk of breast and ovarian cancer are BReast CAncer gene 1(BRCA1) and BReast CAncer gene 2 (BRCA2). The BRCA1 gene is found on chromosome 17q21 and contains 24 exons. This gene has main role to encodes a protein that comprised of 1863 amino acids (5.6 kb)13. BRCA2 gene is found on chromosome 13q12 and contains 27 exons where the biggest exon is number 11 (4.9 kb). The protein that encoded by BRCA2 has 3418 amino acids (10.2 kb). The incidence of ovarian cancer is caused by mutations in exon 13 of BRCA1 and in exon 11 of BRCA2 gene14. The BRCA1 and BRCA2 genes generally operate as tumour suppressors because they code for the proteins implicated in the response to DNA damage. Moreover, they are part of the DNA repair genes that regulate the cell cycle 15.
The significance of the BRCA genes:
BRCA genes contribute to encode of groups of proteins that are necessary for the transcriptional control of DNA synthesis, function as tumour suppressor genes. Therefore, the identification of certain DNA damage, specifically double-stranded breaks, and correct the damage. These mutations may be in several patterns, such as point mutations or deletion/insertion mutations 16. A crucial recombinase enzyme known as RAD51 acts in conjunction with BRCA2 to repair double-stranded DNA breaks and promote homologous recombination. Additionally, exon 11 codes for a structural motif that has eight repeats of “BRC,” through which BRCA2 regulates the function of RAD51. As a result, RAD51 protein and the proteins encoded by the BRCA1/2 tumour suppressor genes work cooperatively to repair DNA breaks, thereby promoting genomic homeostasis17.
The whole genome sequencing analysis for BRCA 1/2
Numerous mutations have been found on the BRCA genes due to their size (large genes). Broadly, it is advisable to carry out a genetic analysis that takes into account both repeated and unusual mutations in the BRCA1 and BRCA2 genes. The typical procedure for the laboratory assessment of BRCA genes comprises extensive genomic rearrangement testing and sequencing. Lately, the mutations in both genes have been studied by using next generation sequencing. A single-site targeted mutation analysis can also be carried out if the patient has a family member who carries a specific mutation18. Clinically, the mutations in BRCA 1 and BRCA 2 were identified in 4–7% of the women that involved in significant studies that used multi-gene analysis. Because it is an expensive and time-consuming method, whole genome sequencing, which involves sequencing all of the DNA’s coding and non-coding regions, is rarely used in clinical practice. As a first-line test for hereditary breast/ovarian cancer, it has not been approved18.
Cancer risk associated with BRCA1/2 mutations
BRCA1 and BRCA2 mutation carriers make up only 0.1–0.2% of the general population. 2-3% of all breast cancer cases have BRCA1 or BRCA2 mutations. Breast and ovarian cancers patients whose have family history in mutations of BRCA genes are more susceptible to mutations at different age stages. Within and between family members, there seems to be variation in the prevalence of pathogenic BRCA mutations and the duration of cancer19. Furthermore, BRCA-associated cancers frequently develop into invasive disease without a precancerous ductal carcinoma in situ component. Consequently, even using mammography to detect breast cancer, the likelihood of doing so is decreased. Several studies have determined that familial ovarian cancer syndromes do not frequently present with mucinous and borderline ovarian carcinomas. Primary cancers of the fallopian tubes and peritoneum are more common in mutation carriers20.
Ovarian cancer risk is increased in people with BRCA2 mutations as well, however it is not as great as it is in people with BRCA1 mutations. Although those who carry the BRCA1 and BRCA2 mutations may experience ovarian cancer at an earlier age, ovarian cancers detected at an older age may also have an underlying mutation13. The presence of abnormalities in the BRCA1 gene has been observed in women with breast cancer only, as well as in cases with both breast and ovarian cancer, at a rate of 15-20% and 40-50%, respectively. A woman with a BRCA1 mutation has a 20% chance of breast cancer after age 40, a 51% risk after age 50, and an 85% risk after age 70. After the age of 70, the likelihood of developing ovarian cancer increases by 40–50%. BRCA2 mutation carriers had a 28 percent increased risk of developing breast cancer after age 50 and an 84 percent increased risk after age 70, and a 4 percent increased risk of developing ovarian cancer after age 50 and a 27 percent increased risk after age 70. In conclusion, having a BRCA1 mutation increases a woman’s lifetime risk of acquiring breast cancer by up to 85% and ovarian cancer by 40 to 50%, whilst having a BRCA2 mutation reduces these risks to 40–45% and 15–30%, respectively13,21.
c-MYC in breast cancer
A group of proto-oncogenes called the Myc genes includes a number of members (L-myc, N-myc and c-myc). The MYC family includes the c-MYC gene, which is found on human chromosome 8. Myc proteins are a subset of the transcription factor family known as “basic/helix-loop-helix/leucine zipper” (HLH-LZ), which are involved in the control of cell growth, differentiation, and apoptosis22.
By a number of processes including transcriptional control of the proximal promoter region, the expression levels of c-MYC are tightly regulated. It has been determined that high levels of c-MYC expression in breast cancer patients cause the formation and progression of the disease22,23.
c-MYC is a crucial regulator of the tumour microenvironment (TME), and it has a role in stromal cell proliferation and angiogenesis, according to growing data (Fig. 3)24. Cancer-associated fibroblasts (CAFs), tumour-associated macrophages (TAMs), vascular endothelial cells (VECs), myeloid-derived suppressor cells (MDSCs), immune cells, inflammatory cells, adipocytes, and myoepithelial cells make up the TME. Extracellular matrix (ECM), extracellular vesicles (EVs), soluble cytokines, and signalling molecules are examples of non-cellular components that play a significant role in TME. Additionally, TME also includes the lymphatic system and blood arteries (Fig. 4)24. The strong connection between TME and tumour growth in breast cancer has been shown in several studies. This may be reliant on the interplay between TME and breast cancer cells that is mediated by c-MYC. Given the critical function of c-MYC in TME and malignant breast cancer25.
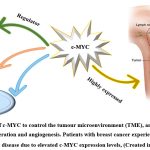 |
Figure 3: The role of c-MYC to control the tumour microenvironment (TME), and implicated in the stromal cell proliferation and angiogenesis. |
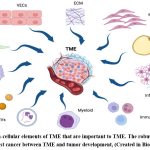 |
Figure 4: Non-cellular elements of TME that are important to TME. The robust relationship found in breast cancer between TME and tumor development, (Created in BioRender.com). |
HER2-positive breast cancer
Several human cancers are the result of some pathogenesis factors such as the human epidermal growth factor receptor (HER) family of receptors. HER family consists of four main groups: HER-1, HER-2, HER-3, and HER-4, also known ErbB1, ErbB2, ErbB3, and ErbB4, respectively. These family of receptors have the fundamental functions, they control the growth of cell, survival, and differentiation through numerous signal transduction methods and participate in cellular proliferation and differentiation26. The cell systems that regulate growth have been the centre of contemporary study into the origins of cancer. This has demonstrated that growth factor receptor alterations have a role in oncogenesis. One of the systems in breast cancer that has been most thoroughly studied is HER family, which plays a significant role in the regulation of healthy growth. HER2 is a significant oncogene in breast cancer and found on the long arm of human chromosome 17 (17q12)27.
The significance of the HER
The HER receptor monomers are present on the cell surface. When ligands bind to the extracellular domains of HER proteins, the intracellular domains undergo dimerization and transphosphorylation. Homo-or heterodimerization leads to the autophosphorylation of tyrosine residues within the cytoplasmic field of the receptors. Thus, it initiates a variation of signalling pathways, principally the mitogen-activated protein kinase (MAPK), phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K), and protein kinase C (PKC) resulting in cell proliferation, survival, differentiation, angiogenesis, and invasion. Heterodimers provide stronger signals than homodimers, and those that contain HER2 have a very high capacity for ligand binding and signalling since HER2 is the family member’s preferred dimerization partner because it occurs in an open conformation26.
The main regulator of cell proliferation and survival known as PI3K/Akt is most effectively stimulated by the HER2-HER3 heterodimer. Furthermore, the mislocalization and quick breakdown of the cell-cycle suppressor p27Kip1 protein, which promotes cell-cycle advancement, are made possible by HER2 dimerization. Furthermore, the mislocalization and quick breakdown of the cell-cycle inhibitor p27Kip1 protein, which promotes cell-cycle advancement, are made possible by HER2 dimerization. Receptors found in other membranes, for example insulin-like growth factor receptor 1, can combine with HER2 to activate it26.
In the HER family, HER2 plays a crucial function by interacting with the other receptors to enhance intracellular signalling12. Studies have connected HER2 gene amplification/receptor overexpression to cancer development despite our poor understanding of how HER2 operates and is controlled. A considerable part of different tumour types exhibit HER2 gene amplification/receptor overexpression, which happens relatively early in the course of disease and is associated with a bad outcome in women with female breast cancer. Making management choices will depend more and more on knowing the HER2 status of patients with breast cancer 28.
Detection of HER2
Despite the development of numerous HER2 testing techniques, approximately 20% of HER2 tests performed today may be inaccurate.. As a result, the College of American Pathologists (CAP) and the American Society of Clinical Oncology (ASCO) have suggested recommendations for HER2 testing to assure accuracy. Immunohistochemistry (IHC) and fluorescence in situ hybridization are the two procedures that are currently authorized for HER2 testing Fluorescence in situ hybridization (FISH)26.
In cases of malignant breast cancer, it is recommended that patients undergo HER2 analysis. And the test should be conducted more than once until the result is confirmed. Initial HER2 testing should be performed on breast cancer tissues employing an IHC technique to detect the presence of the HER2 protein. The first procedure in breast cancer specimens is examine the HER2 by using validated IHC screening for HER2 protein expression29. Depending on the cell membrane staining manner, the grading scheme for HER2 expression is as follows26:
3+: positive HER2 expression, over 30% of malignant tumor cells show homogeneous, strong membrane staining.
2+: equivocal for HER2 protein expression, complete membrane staining that is either faint or inconsistent in intensity, with at least 10% of cells exhibiting a circular distribution.
0 or 1+: negative for HER2 protein expression.
FISH should be used as confirmation on breast cancer tissues with ambiguous IHC results. The results of the HER2-to-CEP17 ratio and gene copy number analysis are interpreted as follows:
Positive HER2 amplification: HER2 gene copy over 6.0 or FISH ratio more than 2.2.
Equivocal HER2 amplification: a HER2 gene copy number of 4.0–6.0 or a FISH ratio of 1.8–2.2.
Negative HER2 amplification: HER2 gene copy less than 4.0 or FISH ratio less than 1.8.
Amplification of the HER2 gene in breast cancer
Since it was discovered that HER2 can cause mammary carcinogenesis both in vitro and in vivo, investigations on the protein have primarily focused on breast cancer. There are prognostic and predictive consequences for the overexpression of HER2 in 15–30% of malignant breast tumors. Approximately from 25 to 50 copies of the HER2 gene and an increase of 40–100 fold in HER2 protein lead to the expression of two million receptors on the tumor’s surface cells, are found in breast tumors30.
It has been demonstrated that even estrogen can activate HER2 signalling by way of the nongenomic activation of the estrogen receptor (ER) outside the nucleus. Breast cancer patients with HER2 gene amplification have lower overall and disease-free survival rates. In a research by Press et al, 704 node-negative breast tumors were examined for HER2 expression, and it was discovered that women whose breast cancers had high overexpression where had a 9.5-fold higher chance of recurrence compared to women whose breast tumors had a normal expression (P = 0.0001)31.
Seshadri et al. discovered that HER2 amplification of thrice or more was related with significantly shorter disease-free survival (P = 0.0027) in their study of 1056 patients with Stages I–III breast cancer32.
Significant correlations between HER2 amplification and the pathologic stage of the disease, the number of axillary nodes that had tumors, the histologic type, and the lack of ER and progesterone receptor (PgR) have also been found. According to available data, HER2 amplification occurs early in the development of human breast tumors. Without any signs of invasive disease, over half of all in situ ductal carcinomas exhibit HER2 amplification, and HER2 status is maintained throughout the development of invasive disease, nodal metastasis, and distant metastases33. Breast cancers with HER2 amplification are more susceptible to some cytotoxic chemotherapeutic treatments, more resistant to some hormonal therapies, and more likely to spread to the brain34.
Conclusion
Breast cancer may be prevented. Two key strategies for preventing breast cancer include lowering risk factors and using chemoprevention. To raise public knowledge of breast cancer, nevertheless, there is still more work to be done. Among high-risk females, just 4.1% are willing to use chemotherapy preventatives. The inability to find the correct method for treating malignant tumors such as breast cancer may be attributed to the genetic nature and its association with a several of factors, and thus the variation in complications between individuals. The Gail method, or International Breast Cancer Intervention Study (IBIS), is one of the most common methods for diagnosing the degree of breast cancer in women. These methods depend on determining age, family history and reproductive ability. However, these methods are inaccurate in their results to determine the severity of the infection in female patients. Due to the development of methods used in studying the genome, it is useful to use genome sequencing in individuals as a biomarker to diagnose the risk of breast cancer. The future goal is to modify the currently used medications and reduce their negative side effects on patients, based on genomic studies.
Acknowledgment
The author would like to thank Professor Sherif Idris Ahmed, Department of Biological Science, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia for his review and revision of the study.
Conflict of Interest
The author declare that I have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Funding Sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
References
- DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66(1):31-42. doi:10.3322/caac.21320
CrossRef - DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J Clin. 2016;66(1):31-42. doi:10.3322/caac.21320
CrossRef - Freitas V, Scaranelo A, Menezes R, Kulkarni S, Hodgson D, Crystal P. Added cancer yield of breast magnetic resonance imaging screening in women with a prior history of chest radiation therapy. Cancer. 2013;119(3):495-503. doi:10.1002/cncr.27771
CrossRef - Majeed W, Aslam B, Javed I, et al. Breast cancer: Major risk factors and recent developments in treatment. Asian Pacific Journal of Cancer Prevention. 2014;15(8):3353-3358. doi:10.7314/APJCP.2014.15.8.3353
CrossRef - Seely JM, Alhassan T. Screening for breast cancer in 2018—what should we be doing today? Current Oncology. 2018;25:S115-S124. doi:10.3747/co.25.3770
CrossRef - Maffini M V., Soto AM, Calabro JM, Ucci AA, Sonnenschein C. The stroma as a crucial target in rat mammary gland carcinogenesis. J Cell Sci. 2004;117(8):1495-1502. doi:10.1242/jcs.01000
CrossRef - Dumars C, Ngyuen JM, Gaultier A, et al. Oncotarget 78343 Www.Impactjournals.Com/Oncotarget Dysregulation of Macrophage Polarization Is Associated with the Metastatic Process in Osteosarcoma. Vol 7.; 2016. www.impactjournals.com/oncotarget/
CrossRef - Basse C, Arock M. The increasing roles of epigenetics in breast cancer: Implications for pathogenicity, biomarkers, prevention and treatment. Int J Cancer. 2015;137(12):2785-2794. doi:10.1002/ijc.29347
CrossRef - Crabtree JS, Miele L. Breast cancer stem cells. Biomedicines. 2018;6(3). doi:10.3390/biomedicines6030077
CrossRef - Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective Identification of Tumorigenic Breast Cancer Cells. www.pnas.orgcgidoi10.1073pnas.0530291100
- Polyak K. Breast cancer: Origins and evolution. Journal of Clinical Investigation. 2007;117(11):3155-3163. doi:10.1172/JCI33295
CrossRef - Sun YS, Zhao Z, Yang ZN, et al. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13(11):1387-1397. doi:10.7150/ijbs.21635
CrossRef - Alacacioglu A, Varol U, Kucukzeybek Y, et al. BRCA genes: BRCA 1 and BRCA 2. JBUON. 2018;23(4):862-866.
- Lane TF, Deng/’^ C, Elson A, Lyu MS, Kozak CA, Leder^ P. Expression of Brcal Is Associated with Terminal Differentiation of Ectodermally and Mesodermally Derived Tissues in Mice.; 1995.
CrossRef - Deng CX. BRCA1: Cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res. 2006;34(5):1416-1426. doi:10.1093/nar/gkl010
CrossRef - Feng Z, Kachnic L, Zhang J, Powell SN, Xia F. DNA damage induces p53-dependent BRCA1 nuclear export. Journal of Biological Chemistry. 2004;279(27):28574-28584. doi:10.1074/jbc.M404137200
CrossRef - Patel KJ, Yu VPCC, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ. Involvement of Brca2 in DNA Repair Tumor Suppressor. The Large Protein It Encodes Does Amino-Terminal Domains in the Transcriptional Regulation. Vol 1.; 1998.
CrossRef - Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387-402. doi:10.1146/annurev.genom.9.081307.164359
CrossRef - Piombino C, Cortesi L, Lambertini M, Punie K, Grandi G, Toss A. Secondary Prevention in Hereditary Breast and/or Ovarian Cancer Syndromes Other Than BRCA. J Oncol. 2020;2020. doi:10.1155/2020/6384190
CrossRef - Tai YC, Domchek S, Parmigiani G, Chen S. Breast cancer risk among male BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2007;99(23):1811-1814. doi:10.1093/jnci/djm203
CrossRef - Guttmacher AE, Collins FS, Wooster R, Weber BL. Breast and Ovarian Cancer. Vol 348.; 2003. http://www.genome.ucsc.edu
CrossRef - Gao F yan, Li X tong, Xu K, Wang R tian, Guan X xiang. c-MYC mediates the crosstalk between breast cancer cells and tumor microenvironment. Cell Communication and Signaling. 2023;21(1). doi:10.1186/s12964-023-01043-1
CrossRef - Liao DJ, Dickson RB. C-Myc in Breast Cancer. Vol 7.; 2000. http://www.endocrinology.org
CrossRef - Meškytė EM, Keskas S, Ciribilli Y. Myc as a multifaceted regulator of tumor microenvironment leading to metastasis. Int J Mol Sci. 2020;21(20):1-29. doi:10.3390/ijms21207710
CrossRef - Tyan SW, Kuo WH, Huang CK, et al. Breast cancer cells induce cancer-associated fibroblasts to secrete hepatocyte growth factor to enhance breast tumorigenesis. PLoS One. 2011;6(1). doi:10.1371/journal.pone.0015313
CrossRef - Iqbal N, Iqbal N. Human Epidermal Growth Factor Receptor 2 (HER2) in Cancers: Overexpression and Therapeutic Implications. Mol Biol Int. 2014;2014:1-9. doi:10.1155/2014/852748
CrossRef - Wolff AC, Elizabeth M, Hammond H, et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. Vol 131.; 2007. http://arpa.allenpress.com
CrossRef - Harbeck N. Neoadjuvant treatment of HER2-positive breast cancer: should therapy differ based on hormone receptor status? Ther Adv Med Oncol. 2018;10. doi:10.1177/1758835918782356
CrossRef - Wolff AC, Elizabeth M, Hammond H, et al. Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. Vol 131.; 2007. http://arpa.allenpress.com
CrossRef - Kallioniemi OP, Kallioniemi A, Kurisut W, et al. ERBB2 Amplification in Breast Cancer Analyzed by Fluorescence in Situ Hybridization. Vol 89.; 1992. https://www.pnas.org
CrossRef - Press MF, Pike MC, Chazin VR, et al. Her-2//Z£M Expression in Node-Negative Breast Cancer: Direct Tissue Quantitation by Computerized Image Analysis and Association of Overexpression with Increased Risk of Recurrent Disease1. http://aacrjournals.org/cancerres/article-pdf/53/20/4960/2452198/cr0530204960.pdf
- Seshadri R, Firgaira FA, Horsfall DJ, McCaul K, Setlur V, Kitchen P. Clinical significance of HER-2/neu oncogene amplification in primary breast cancer. Journal of Clinical Oncology. 1993;11(10):1936-1942. doi:10.1200/JCO.1993.11.10.1936
CrossRef - Stewart RL, Caron JE, Gulbahce EH, Factor RE, Geiersbach KB, Downs-Kelly E. HER2 immunohistochemical and fluorescence in situ hybridization discordances in invasive breast carcinoma with micropapillary features. Modern Pathology. 2017;30(11):1561-1566. doi:10.1038/modpathol.2017.65
CrossRef - Gabos Z, Sinha R, Hanson J, et al. Prognostic significance of human epidermal growth factor receptor positivity for the development of brain metastasis after newly diagnosed breast cancer. Journal of Clinical Oncology. 2006;24(36):5658-5663. doi:10.1200/JCO.2006.07.0250
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.






