How to Cite | Publication History | PlumX Article Matrix
Geethu Suresh* , R. Ragunathan
, R. Ragunathan and Jesteena Johney
and Jesteena Johney
Department of Biotechnology, Centre for Bioscience and Nanoscience Research, Eachanari, Coimbatore, Tamil Nadu 641021 India.
Corresponding Author E-mail:ammuge@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/3186
ABSTRACT: The mounting concerns over the usage of enormous amount of wood and perilous chemicals in paper industry have led the researchers to develop ecofriendly green technologies for pulp and paper production. Lignocellulosic agro-waste can be an excellent alternative to the wood in papermaking. White rot fungi with ligninolytic properties can be effectively used for pretreatment of agro-wastes for pulp making. This study isolates and identifies previously unexplored strains of white rot fungi from Western Ghats of Kerala, India which can be used for developing biological systems for lignocellulosic treatment in pulp and paper production. Ten isolates were identified using molecular techniques and screened for their extracellular enzyme activities, including laccase, Manganese peroxidase and Lignin peroxidase. Among the 10 isolates, Trametes versicolor (Laccase activity 31.79 U/ml, Manganese peroxidase (MnP) 42.336 U/ml and Lignin peroxidase (LiP) - 50.65U/ml, Favolus teniculus (Laccase - 41.54 U/ml, MnP - 44.07 U/ml &LiP - 30.54), Coriolopsis byrsina (Laccase - 42.56 U/ml, MnP - 43.54 U/ml and LiP - 25.14 U/ml) Lenzitus betulina (Laccase - 37.15 U/ml, MnP - 38.97 U/ml and LiP - 30.43 U/ml) exhibited highly promising lignolytic enzymatic system. The study’s findings may provide a better eco-friendly substitute for conventional chemical treatments in various industrial applications.
KEYWORDS: Coriolospsis byrsina; Favolus teniculus; Lignin; laccase; Lignin peroxidase; Lenzitus betulina; Manganese peroxidase
Download this article as:| Copy the following to cite this article: Suresh G, Ragunathan R, Johney J. Screening and Molecular Identification of Potential Lignolytic White Rot Fungi Isolated from Western Ghats. Biotech Res Asia 2023;20(4). |
| Copy the following to cite this URL: Suresh G, Ragunathan R, Johney J. Screening and Molecular Identification of Potential Lignolytic White Rot Fungi Isolated from Western Ghats. Biotech Res Asia 2023;20(4). Available from: https://bit.ly/3RGFd5Q |
Introduction
Lignocellulosic waste generated from forestry and agricultural practices constitutes a significant portion of municipal solid wastes and causes substantial environmental pollution. The escalating demands for wood resources in the paper making industry and the use of hazardous chemicals in paper production pose additional threat to the environment. Fungi, especially white rot fungi are recognized as the major organisms for lignocellulosic degradation 1,28. White rot fungi are the major organisms which are capable of effective lignin mineralization. Efficient removal of lignin from lignocellulosic wastes makes them a better alternative for wood in paper making.
Pretreatment of lignocellulosic wastes using microorganisms and their enzymatic machinery can efficiently break down lignin and alter lignocellulose. White rot fungi is regarded as the greatest promising microorganism which can mineralize lignin to CO2 and water in pure culture3. White rot fungi produces variety of extracellular lignolytic enzymes such as Laccase(EC1.10.3.2) and peroxide reductases or peroxidases which includes Manganese peroxidase- MnP (EC 1.11.1.13), Lignin peroxidase- LiP (EC1.11.1.14) and versatile peroxidase (EC1.11.1.16) 1,4). Lignin modifying enzymes are the product of secondary metabolism of white rot fungi.
In recent years, the studies on lignin modifying white rot fungi have been increased in search of efficient lignolytic machinery to utilize lignocellulosic biomass more effectively. The lignolytic properties of white rot fungi can be employed in many aspects of lignolytic biomass pretreatment such as Bioethanol production9, pulp and paper making 11,12 and bioremediation of xenobiotic compounds13. The capability of white rot fungi to generate lignolytic enzymes is the key focus of these studies. Various white rot fungi have been reported to exhibit lignin degrading ability. Previous studies have mainly focused on Phenerochete chryosporium, Cereporiopsis subvermispora, Trametes versicolor, pleorotus ostreatus5,6,7,8 as the widely studied white rot fungi.
However, the identification of alternative and unexplored strains with potential lignolytic abilities is critical for the development of feasible biotechnological systems for various industrial applications and proposing a more eco-friendly substitute for conventional chemical treatments. Western Ghats are the hotspots of biodiversity with varied flora and fauna. In this study fruit bodies have been collected from Western Ghats of Kerala and it is expected to provide unexplored candidates with potential lignolytic machinery for various biotechnological applications.
Thus, the present study aims to (i) isolate white rot fungi from Western Ghats, molecular identification of the strain ii) screening of this isolates for its ability to degrade lignin in order to find unexplored strains with high lignin degrading system.
Materials and Methods
Collection and culturing of fungal samples
The samples including fruiting bodies from lignocellulosic rich litter piles and decayed woods from Vellanikkara area of (10.5449° N, 76.2864° E) Western Ghats were collected in sterile zip lock polythene bags and brought to the laboratory for further analysis.
 |
Figure 1: Location map of site of sample collection |
Total number of 10 fruiting bodies were collected. The collected samples were cultured in MGYP broth (3 g Malt extract, 10g Glucose, 03g Yeast extract, 05 g Peptone per litre of distilled water) and Malt Extract Agar medium. The fruiting bodies were cut into pieces and surface sterilized with 70% ethanol inoculated in 25 ml of MGYP medium. It was then kept under room temperature for 4 to 6 days14. The pure culture was then maintained by subculturing in 2%Malt Extract Agar medium.
Molecular identification of fungal samples
DNA isolation was done by CTAB method.14. The Isolated DNA was then amplified by using I8S rRNA primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′); ITS4: (5′-TCCTCCGCTTATTGATATGC-3′). The reaction mixture consisted isolated DNA-2µl, ITS primers-1µl each, pcr master mix – 10µl, and sterile water -8µl. PCR cycles have been carried out with an initial denaturation temperature of 950C for 2 minutes and the other conditions were: denaturation – 950C – 1 minute, annealing – 550 C -1 minute, extension – 720C – 1 minute and final extension at 720C for 5 minutes. The total number of cycle was 30. The amplicon obtained was electrophoresed using 1.2% agarose gel and imaged in a gel documented. The product was then purified using purification column and sequenced (Chromus Biotech, Banglore). The sequence obtained was deposited to NCBI databank to get the accession number and it was analyzed using BLAST, a NCBI tool. The sequences were compared with the similar sequences available in the Genbank by Multiple sequence alignment using the tool ClustalW. The aligned data was used to construct phylogenetic tree using the software MEGA7.
Primary screening
Dye decolourization method using Methylene Blue:
The fungal culture was inoculated in malt extract agar plates containing 0.01% Methyelene blue as described by Sasikumar et al. with slight modification24 and incubated at 25ºC for two weeks. A clear zone formation around the colony will indicate the presence of enzyme Lignin peroxidases.
Guaiacol agar plate assay
Malt extract agar plates with 0.01% guaiacol were prepared and the fungal culture was inoculated. It was then incubated at 25ºc for two weeks. Positive reaction will form brown color which indicates the presence of Laccase enzyme15.
Secondary screening by quantitative Enzyme assays
Fungal mycelium discs of 6mm were cultured in 20 ml of Mineral Salt medium and incubated for 12 days (composition g/L: Glucose-10g/L;Pottasium dihydrogen orthophosphate -2 g/L;Magnesium sulphate hepta hydrate -0.5 g/L;Calcium chloride-0.1 g/L; Ammonium tartrate -0.2 g/L. Trace element solutioncomprised of (mg/L) Ferrous sulfate-12;Manganese sulphate- 3;Zinc sulfate- 3;Cobalt sulfate hepta hydrate -1;Ammonium molybdate -1(pH. 5.0)16. After incubation the culture was filtered using Whatman No1 filter paper. Around 5 ml of supernatant was used to analyze the extracellular enzymes. All the experiments were done in triplicates.
Laccase (E.C.1.10.3.2)
For determining the activity of laccase enzyme, guaiacol(2 mM) was used as the substrate. Reaction mixture was prepared using Guaiacol(2 mM) in sodium acetate buffer (pH 4.5). 100 µl of enzyme was added and the mixture was incubated at 30oC. Absorbance was taken at 450 nm. The quantity of laccase enzyme needed to oxidize 1µm of guaiacol per minute15 is regarded as one unit enzyme activity. The extinction coefficient of guaiacol at 450 nm is 12,100 M-1cm-1
Lignin peroxidase(EC.1.11.1.14)
LiP enzyme activity was analyzed according to Tien & kirk method (1989)25which relies on the rate of oxidation of veratryl alcohol. The reaction mixture constitutes 250 µl of sodium tartarate buffer, 100 µl of veratryl alcohol(2 mM) and 100 µl of supernatant. The reaction was commenced by the adding of 100 µl H2O2. The absorbance was taken at 310 nm. One unit of enzyme activity is expressed as the quantity of enzyme required to oxidize 1µm of veratryl alcohol/minute.
Manganese peroxidase ( E.C.1.11.1.13)
Manganese peroxidase assay was carried out based on the oxidation of guaiacol. The reaction mixture consists of 100 µl of sodium tartarate buffer (pH 5.0) ,10 µl of guaiacol, 10 µl of MnSO4 and 100 µl of enzyme. Addition of 10 µl of H2O2 commenced the reaction and absorbance was measured at 465 nm15.
Result and Discussion
White rot fungi which produces ligninolytic enzymes are important biological tools with possible application in wide variety of fields. The search of strong lignin degrading enzymatic machinery has always been an interesting topic among researchers owing to its potential applications. In this study 10 samples from Western Ghats have been isolated and identified.
The biodegradation ability of saprophytic basidiomycetes is closely connected with the production of extracellular enzymes, among them, Laccase, Lip, MnP plays important roles17. Some wood degrading fungi produces all these three enzymes, while others produces only one or two of these enzymes.6. These enzymes are not only involved in lignin degradation as observed in the delignification of natural lignocellulosic materials, but also actively degrade many recalcitrant compounds including dyes18.
Primary screening for strains producing lignolytic enzymes
The initial screening strategy used for detecting the capability of fungal strains to produce lignin degrading enzymes was the dye decolourizing technque18. The fungi capable of producing lignolytic enzymes which decolorizes the azo dye Methylene blue were screened. During dye degradation, the azo linkage is cleaved by oxidation by lignolytic enzymes like laccase, MnP, LiP produced by the white rot fungi19. Lignolytic enzymes are oxidative in nature and hence are able to decolorize basic dyes by reduction.
In the study, all the isolates were positively responded to the dye decolorization assay using methylene blue. Clear zones were formed around the colonies indicating the presence of lignin modifying enzymes. (Figure 2). This suggests all the isolates are able to produce any of the oxidative lignolytic enzymes. In line with the studies by Sasikumar et al. 24, the decolorization of methylene blue to clear zones can be used as indicative of production of Lignin peroxidase enzyme.24. This suggests all the ten isolates are capable of producing Lignin peroxidase enzyme.
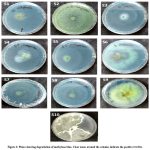 |
Figure 2: Plates showing degradation of methylene blue. Clear zones around the colonies indicate the positive reaction. |
The ability of fungi to produce lignolytic enzymes can be verified by its ability to oxidize phenolic compounds like guaiacol and syringaldazine21. 10 samples were tested for the ability to oxidize guaiacol, to screen potential isolates which degrade lignin.
All the isolates formed reddish brown color zone around the colony, indicating the presence of laccase enzyme. (figure2). Production of reddish brown zone is due to the oxidation of guaiacol by laccase enzyme15. The similar results reported by Anuja&Aggarwal et al.15 and Dhoib et al.18in well studied white rot fungi such as Phenerochaete chryosporium, Trametes versicolor, Pleurotus ostreatus, Ganoderma lucidum.
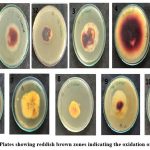 |
Figure 3: Plates showing reddish brown zones indicating the oxidation of guaiacol |
Table 1: Primary screening of isolates for detecting the presence of lignolytic enzymes.
| Isolate | Name of the strain | Primary screening | |
| Methylene blue test – zone of clearance
(mm) |
Guaiacol plate test
-reddish brown zone (mm) |
||
| CBNR 1 | Trametes versicolor (OL636386) | 3 | 4 |
| CBNR 2 | Coriolopsis byrsina (OL587502) | 5 | 2 |
| CBNR3 | Lenzitus betulinus (OM424452) | 7 | 4 |
| CBNR 4 | PhenerochaeteSp (OL636387) | 6 | 2 |
| CBNR 5 | Phenerochaete avellanea (ON945561) | 5 | 3 |
| CBNR 6 | Ganoderma austral (OL636391) | 10 | 8 |
| CBNR 7 | Rickenella fibula (OL636388) | 12 | 6 |
| CBNR 8 | Favolus teniculus (OL636390) | 12 | 2 |
| CBNR 9 | Coriolopsis byrsina (ON945562) | 12 | 2 |
| CBNR 10 | Coriolopsis byrsina (OL539546) | 12 | 6 |
Molecular identification of fungal strains
Genetic characterization using 18S rRNA has been considered as a sensitive tool to identify the fungal taxa. The Internal Transcribed Spacer (ITS) region is the suggested barcode for fungi since it has got good species resolution which covers broad spectrum of fungi. Banos et al. 23 describes the efficacy of using ITS region for identification of fungi in comparison to other marker genes. The molecular studies of lignin degrading white rot fungi have been reported in studies of Roseline et al. 29 and Badr El-Din et al. 11.
The strains having maximum homology were compared and aligned with ClustalW. The accession numbers obtained are OM424452, OL636391, OL636390, OL636388, OL636387, OL636386, OL587508, OL539546, ON945561 and ON945562. The dendrogram was constructed comparing fungal 18S rRNA gene sequences obtained with the similar sequences available in the NCBI Genbank.(figure 4 A,B,C,D).
 |
Figure 4: Dendrogram showing 18s rRNA sequencing and sequence similarities of the isolates with related genus. |
Quantitative screening for Laccase, LiP and MnP
The isolates, after primary screening were further quantitatively analyzed for their ability to produce Laccase, Lip And MnP enzymes. Lignin modifying activity of white rot fungi are predominantly associated with the secretion of enzymes such as Laccase, lignin peroxidase and manganese peroxidase.
Enzyme production was carried out under static flask conditions using basal medium. Crude enzyme was filtered out and checked for the enzyme activity. In this study, all 10 strains exhibited lignolytic activities to varying degrees. Also, white rot fungi which are hardly exploited and studied for its lignolytic potentials have been identified. Favolus teniculus(OL636390), Coriolopsis byrsina(OL539546), Lenzitus betulina(OM424452) were positively responded to dye decolorization test and shown good Laccase, MnP and LiP activities on respective enzyme assays, suggesting they have lignin modifying abilities.
Trametes versicolor (Laccase activity 31.79 U/ml, MnP- 42.336 U/ml andLiP- 50.65), Favolus teniculus (Laccase activity 41.54 U/ml, MnP- 44.07 U/ml andLiP- 30.54 U/ml), Coriolopsis byrsina(Laccase activity 42.56 U/ml, MnP- 43.54 U/ml and LiP- 25.14) Lenzitus betulinus (Laccase activity 37.15U/ml, MnP- 38.97 U/ml &LiP – 30.43U/ml) were find to have promising lignolytic enzymatic system. (figure 5). Aydınoğlu, 27 studied optimization of production of laccase from Trametes versicolor and reports laccase activity of 30.28 /g which is similar to our studies. Yuliana et al. 30 reports Laccase from Trametes versicolor with specific activity of 3.78 U/mg and purified enzyme shows the specific activity of 10.96 U/mg . However, the studies on enzymatic systems of other fungi Favolus teniculus, Coriolopsis byrsina, Lenzitus betulinus are scarce.
Laccase activity of these isolates ranged from 3.9U/ml to 45.75U/ml with the highest being produced by Ganoderma austral. Jing si et al. 31 reports the specific laccase activity of laccase enzyme isolated from Ganoderma australe as 22.214 U/mg and a putification fold of 23.989. MnP activity spanned 5.79 U/ml to 44.07 U/ml with the highest producer being Favolus teniculus. The enzyme activity of Favolus teniculus is being reported for the first time. Lignin peroxidase activity having a range of 13.1 U/ml to 50.65 U/ml where Trametes versicolor being the highest producer. Pooja singh et al. 32 studied the lignolytic enzyme activities of Trametes versicolor on oil palm biomass and reports the lignin peroxidase activity of 42.56 U/L.
While dye decolourization study, the isolates Favolus teniculus(OL636390), Coriolopsis byrsina(OL539546) and Lenzitus betulina(OM424452) were able to decolorize the dye methylene blue. This indicates the presence of extracellular oxidative enzymes produced by them, as reported by Dastidar et al10.Similarly, all the three isolates were able to oxidize Guaiacol by forming a reddish brown zone indicating the presence of laccase enzyme produced by them. Further, quantitative enzyme assays has given a good picture regarding the lignolytic enzyme production by the three isolates. (Table 2).
Hataka et al.6 studied lignolytic enzymes of white rot fungi and reported ligninolytic enzymes of Phenerochete chryosporium, Cereporiopsis subvermispora, Pleurotus ostreatus. Ramanaiah Illuri et al.33 reports the lignolytic enzymes of basidiomycetes especially Pleurotus sp. Kirk et al.5 detailed the mechanisms of enzyme production in Phenerochete chryosporium, Cereporiopsis subvermispora. The present result correlates with the previous reports and justifies the isolates have the potential to degrade lignocellulose effectively.
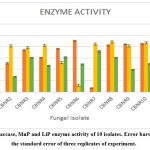 |
Figure 5: Laccase, MnP and LiP enzyme activity of 10 isolates. Error bars represent the standard error of three replicates of experiment. |
Table 2. Molecular identification and screening of isolates for enzymatic activity
| Isolate | Name of the strain | Primary screening | Enzyme assay( U/ml) | |||
| Methylene blue test | Guaiacol plate test | Laccase | LiP | MnP | ||
| CBNR 1 | Trametes versicolor (OL636386) | +ve | +ve | 31.37 | 50.65 | 42.33 |
| CBNR 2 | Coriolopsis byrsina (OL587502) | +ve | +ve | 26.47 | 13.60 | 40.60 |
| CBNR3 | Lenzitus betulinus (OM424452) | +ve | +ve | 37.46 | 30.43 | 38.97 |
| CBNR 4 | PhenerochaeteSp (OL636387) | +ve | +ve | 30.16 | 23.55 | 41.09 |
| CBNR 5 | Phenerochaete avellanea (ON945561) | +ve | +ve | 38.37 | 30.87 | 25.81 |
| CBNR 6 | Ganoderma austral (OL636391) | +ve | +ve | 45.75 | 24.53 | 5.79 |
| CBNR 7 | Rickenella fibula (OL636388) | +ve | +ve | 3.99 | 36.20 | 42.33 |
| CBNR 8 | Favolus teniculus (OL636390) | +ve | +ve | 39.39 | 30.54 | 44.07 |
| CBNR 9 | Coriolpsis byrsina (ON945562) | +ve | +ve | 42.53 | 20.07 | 40.73 |
| CBNR 10 | Coriolopsis byrsina (OL539546) | +ve | +ve | 42.94 | 25.14 | 43.54 |
The potential industrial applications of white rot fungi in lignocellulosic waste management are numerous, and our study provides additional evidence of the effectiveness using such fungi to degrade lignin. Moreover, identifying new strains of white rot fungi could provide more sustainable and eco-friendly alternative to traditional chemical treatments for lignocellulosic waste. Optimizing the growth conditions of fungi through further investigations can enhance their lignolytic activity, thus leading the process efficient and cost-effective for industrial applications.
Conclusion
White rot fungi are the major group of basidiomycete that has adept capacity for efficient mineralization of lignin. Lignolytic enzymes of these white rot fungi have several industrial and biotechnological applications. In the present study 10 potential lignolytic enzyme producing White rot fungi were isolated, screened and molecular biologically identified using 18s RNA. Our study provides evidence of potential of white rot fungi as a sustainable alternative for lignocellulosic treatments. The isolation and screening of new strains such as those identified in our study (Favolus teniculus, Coriolopsis byrsina and Lenzitus betulina) could lead to the development of more effective and efficient biotechnological systems for various industrial applications.
Conflict of Interest
There is no conflict of interest.
Funding Sources
There is no funding sources.
References
- Kent Kirk, Farrell Enzymatic combustin: microbial degradation of lignin. Ann. Review of microbiology. 1987; 41:465-501.https://doi.org/10.1146/annurev.mi.41.100187
CrossRef - Dashtban M, Schraft H, Syed TA, Qin W. Fungal biodegradation and enzymatic modification of lignin. Int J Biochem Mol Biol. 2010;1(1):36-50.
- Akhtar, M., Blanchette, R.A., Kent Kirk, T. Fungal delignification and biomechanical pulping of wood. Biotechnology in the Pulp and Paper Industry. in Biochemical Engineering/Biotech. 1997; vol 57. Springer, Berlin, Heidelberg. https://doi.org/10.1007/BFb0102074
CrossRef - A.M. Rodrigues, P. Pinto. Effect of enzyme extracts isolated from white-rot fungi on chemical composition and in vitro digestibility of wheat straw. Anim. feed sci. tech. 2008; 141: 326-338 https://doi.org/10.1016/j.anifeedsci.2007.06.015.
CrossRef - Kent Kirk, SukiCroan, Ming Tien .Production of multiple ligninases by Phanerochaete chrysosporium: effect of selected growth conditions and use of a mutant strain. Enz. micro.tech.1986; 8(1):27-32. https://doi.org/10.1016/0141-0229(86)90006-2.
CrossRef - Chi, Yujie&Hatakka, Annele&Maijala, Pekka. Can co-culturing of two white-rot fungi increase lignin degradation and the production of lignin-degrading enzymes. Biodeterioration &Biodegradation. 2007; 59. 32-39.https://doi.org/10.1016/j.ibiod.2006.06.025
CrossRef - S Archibald, R Bourbonnais, L Jurasek, M.G Paice, I.D Reid. Kraft pulp bleaching and delignification by Trametes versicolor. J.Bioteh 1997; 53 (2-3) 215-236 https://doi.org/10.1016/S0168-1656(97)01675-1.
CrossRef - OrlyArdon, Zohar Kerem, and Yitzhak Hadar. Enhancement of lignin degradation and laccase activity in Pleurotus ostreatusby cotton stalk extract, J.Microbiol. 1998; 44(7): 676-680. https://doi.org/10.1139/w98-054
CrossRef - Caixia Wan, Yebo Li. Fungal pretreatment of lignocellulosic biomass. Adv. 2012; 30(6)1447-1457. https://doi.org/10.1016/j.biotechadv.2012.03.003
CrossRef - Dastidar MG, Simha AN, Koushik AB. Bioremediation of industrial effluents by ligninolytic microbes. Int J Dev Res. 2018; 08:22419-22424.
- Badr El-Din, Said & Kheiralla, Zeinab&Malek, Saad& Hussein, Douaa. Selection of Fungal Isolates for Biopulping of Rice Straw. 2013; 8. 10.15376/biores.8.4.4969-4980. DOI:10.15376/biores.8.4.4969-4980.
CrossRef - Bajpai, P.K., Kondo, R. Pulp Bleaching with White Rot Fungi and Their Enzymes In: Biotechnology for Environmental Protection in the Pulp and Paper Industry. Springer, Berlin, Heidelberg,1999; https://doi.org/10.1007/978-3-642-60136-1_5
CrossRef - Kaur, Harsimran, ShammiKapoor, and GaganjyotKaur. Application of ligninolytic potentials of a white-rot fungus Ganodermalucidum for degradation of lindane. monitoring and assessment2016; 188.10 : 1-10.
CrossRef - Johney, Jesteena &Sellappan, s. RadhaiSri &Ragunathan, R. Extraction of Chitin and Chitosan from Wild Type PleurotusSpp and its Potential Application – Innovative Approach. Pure Appl. Microbiol. 2018; 12. 1631-1640. 10.22207/JPAM.12.3.70. DOI:10.22207/JPAM.12.3.70
CrossRef - Sharma, Anuja&Aggarwal, Neeraj&Anitayadav,. Isolation and Screening of Lignolytic Fungi from Various Ecological Niches. 2017; J. Microbiol. Res DOI:10.13189/ujmr.2017.050202
CrossRef - Nikki Agrwal, Preethisharma. Ligninolytic Enzyme Production by White Rot Fungi Podoscypha elegans Strain FTG4. J.Curr.Microbiol.App.Sci .2017; 6(5): 2757-2764. DOI https://doi.org/10.20546/ijcmas.2017.605.309.
CrossRef - Eichlerová I, Baldrian P.Ligninolytic Enzyme Production and Decolorization Capacity of Synthetic Dyes by Saprotrophic White Rot, Brown Rot, and Litter Decomposing Basidiomycetes. Fungi (Basel). 2020; Nov 19;6(4):301.doi: 10.3390/jof6040301.
CrossRef - Dhouib, Abdelhafidh & Hamza, Manel & Zouari-Mechichi, Hela&Mechichi, Tahar&Hmidi, Rafik&Labat, Marc & Martinez, Maria Jesus &Sayadi, Sami. Screening for Ligninolytic Enzyme Production by Diverse Fungi from Tunisia. World J. Microbiol .Biotech. 2005; 21. 1415-1423. DOI:1007/s11274-005-5774-z
CrossRef - Madhumita Ghosh Dastidar, AkshayaSimha, N., Ananth Koushik, B., Aradhya, D., Harishmitha, N., Nayana, S., Nidhish, P., Shakthi Vignesh, S., Siri, G. and Tavisha, V. K..Bioremediation of industrial effluents by ligninolytic microbes. J. Dev. Res. 2018; 8, (08), 22419-2242
- Mamatha Pingili, Shailaja Raj Marla. Isolation and screening of lignin degrading fungi from degraded fruit litter.J.Curr.Microbiol.App.Sci. 2017;6(12): 2200-2206 https://doi.org/10.20546/ijcmas.2017.612.252
CrossRef - Erden E, Ucar MC, Gezer T, Pazarlioglu NK. Screening for ligninolytic enzymes from autochthonous fungi and applications for decolorization of Remazole Marine Blue. Braz J Microbiol. 2009 ;Apr;40(2):346-53. doi: 10.1590/S1517-838220090002000026.
CrossRef - Miki Y, Ichinose H, Wariishi H. Molecular characterization of lignin peroxidase from the white-rot basidiomycete Trametes cervina: a novel fungal peroxidase. FEMS MicrobiolLett. 2010; 304(1):39-46. DOI: 1111/j.1574-6968.2009.01880.x
CrossRef - Banos, S., Lentendu, G., Kopf, A. A comprehensive fungi-specific 18S rRNA gene sequence primer toolkit suited for diverse research issues and sequencing platforms. BMC Microbiol. 2018; 18-190. https://doi.org/10.1186/s12866-018-1331-4
CrossRef - Sasikumar, V., Priya, V., Shankar, C. S., &Sekar, S. D. Isolation and preliminary screening of lignin degrading microbes. Acad.Ind. Res. 2014; 3(6), 291-294.
- Farrell, R. L., Murtagh, K. E., Tien, M., Mozuch, M. D., & Kirk, T. K.. Physical and enzymatic properties of lignin peroxidase isoenzymes from Phanerochaete chrysosporium. Enz.Micro.Tech.1989;11(6), 322–328.doi:10.1016/0141-0229(89)90014-8
CrossRef - Changwei Zhu, GuangwenBao& Shun Huang Optimization of laccase production in the white-rot fungus Pleurotus ostreatus (ACCC 52857) induced through yeast extract and copper, Biotechnology & Biotechnological Equipment,2016; 30:2, 270-276, https://doi.org/10.1080/13102818.2015.1135081
CrossRef - Aydınoğlu, T., Sargın, S. Production of laccase from Trametes versicolorby solid-state fermentation using olive leaves as a phenolic substrate. Bioprocess Biosyst. Eng 2013;36, 215–222 ,. https://doi.org/10.1007/s00449-012-0777-2
CrossRef - Herman Suryadi, Jessica J. Judono, Merianda R. Putri, Alma D. Eclessia, Jiihan M. Ulhaq, Dinar N. Agustina, Triyani Sumiati. Biodelignification of lignocellulose using ligninolytic enzymes from white-rot fungi, Heliyon, 2022; Volume 8, Issue 2. https://doi.org/10.1016/j.heliyon.2022.e08865.
CrossRef - Jebapriya, Roseline & Gnanadoss, J, Joel. Screening and Molecular Characterization of white rot fungi capable of laccase production and dye decolourization. J.Life Science. Pharma Res,2014; 4. 12-20.
- Yuliana, R. Ashifa Putri, I. Hanidah, E. Mardawati and H. Tjaturina. Purification and Characterization Laccase from Trametes versicolor (L.) Lloyd in Submerged Fermentation. Pak Jour of Biological Sciences, 2022; 25: 1077-1084. DOI: 10.3923/pjbs.2022.1077.1084.
CrossRef
- Jing Si, Yi Wu, Hong-Fei Ma, Yong-Jia Cao, Yi-Fei Sun, Bao-Kai Cui, Selection of a pH- and temperature-stable laccase from Ganoderma australe and its application for bioremediation of textile dyes, Jour. of Env. Management, 2021;Volume 299,https://doi.org/10.1016/j.jenvman.2021.113619.
CrossRef
- Singh, Pooja & Sulaiman, Othman & Hashim, Rokiah & Peng, Leh Cheu & Singh, Rajeev. Evaluating biopulping as an alternative application on oil palm trunk using the white-rot fungus Trametes versicolor. International Biodeterioration & Biodegradation,2013;82. 96–103. 10.1016/j.ibiod.2012.12.016.
CrossRef
- Ramanaiah Illuri, M. Kumar, M. Eyini, V. Veeramanikandan, Khalid S Almaary, Yahya B. Elbadawi, M.A. Biraqdar, P. Balaji. Production, partial purification and characterization of ligninolytic enzymes from selected basidiomycetes mushroom fungi. Saudi Journal of Bio.Sci,2021;7207-7218 https://doi.org/10.1016/j.sjbs.2021.08.026.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





