How to Cite | Publication History | PlumX Article Matrix
Restriction digestion of EcoRI in E. coli RY13 plasmid DNA
C.S. Senthilkumar, Tahir M. Malla, Sameena Akhter, Aadil Beg And N. Ganesh
Department of Research, Jawaharlal Nehru Cancer Hospital and Research Centre, Bhopal India.
ABSTRACT: An attempt was made to support the evidenced research of restriction enzyme EcoRI digests plasmid DNA of a significant genetic strain E.coli RY13 by targeting their specific sequence GAATTC. E.coli RY13 was harvested with special antibiotics in terrific broth and plasmid DNA was isolated and digested with EcoRI, then electrophoresed in 0.8% Agarose gel. EcoRI cleaved plasmid DNA into fragments similarly in size that confirmed the presence of target sequence in the E. coli RY13 genome.
KEYWORDS:
Restriction digestion; plasmid DNA; EcoRI, agarose gel; electrophoresis.
Download this article as:| Copy the following to cite this article: Senthilkumar C.S ,Malla T.M , Akhter S, Beg A ,Ganesh A. Restriction digestion of EcoRI in E. coli RY13 plasmid DNA.Biosci Biotechnol Res Asia 2008;5(2). |
Introduction
DNA analysis is a hallmark invention in molecular biology untied our basic understanding strategies behind the art of gene cloning, human genome and variome projects. The foremost aspect of gene cloning involved in the creation of new combination of genetic material from different origin. Definitely, the construction of recombinant DNA molecule requires vector as a vehicle to carry a foreign DNA to the host organisms for gene expression. Vector should possess specific characteristics such as a replicon, resistance to antibiotics or auxotrophic markers and unique cleavage sites for restriction enzymes for foreign DNA insertion. The concept of plasmid as a vector inborned in the concept of particulate determinants of inheritance. Plasmids are circular, autonomously replicating, extra chromosomal DNA molecules found in most gram positive and gram negative organisms and also in some yeast, but not in higher eukaryotes1. Plasmids of E. coli were designated as one of the efficient cloning vectors2. Techniques for cleaving DNA molecules in to discrete fragments by use of specific enzymes virtually known after the proposal of restriction and modification in bacterial viruses – Phage λ3. Further, it was revealed that restrictions were caused by an enzymatic cleavage of incorporating DNA with restriction endonucleases and modification leads to DNA methylation4. Restriction enzymes are recognized as molecular scissors and scalpels in recombinant DNA technology. They are applied in the preparation of recombinant molecules and act as supportive system for the analysis of sequence specific DNA – Protein interactions5. Restriction system consist of two enzymatic elements namely a nuclease and methylase; both are encoded by bacterial chromosomes or by phage or plasmid. Type I, II, III are three kinds of endonucleases that play a key role in cleavage of DNA molecules6. Escherichia coli RI (EcoRI) endonuclease is a renowned restriction enzyme that recognizes the symmetrical palindromic hexanucleotide sequence GAATTC on duplex DNA and cleaves each strand between G and A residues7. Physical and catalytic properties of EcoRI restriction endonuclease also studied by several groups and different purification protocols have been described 8 – 10. In addition to the natural overproducer of EcoRI, E. coli RY13, genetically modified overproducing strains were also used to produce the enzyme11.
Materials and Methods
Harvesting and Purification of Bacterial strain
A significant genetic bacterial strain E.coli RY13 maintained in Genetic engineering laboratory, Centre for Biotechnology was harvested in 20 mL terrific broth containing antibiotic ampicillin grown overnight in an incubator shaker at 37ºC then inoculated and incubated with chloramphenicol for 2 hrs. Cultures were centrifuged at 4000 rpm for 5 mins and pellets obtained were subjected to plasmid DNA isolation.
Plasmid DNA isolation and Restriction digestion
Plasmid DNA was isolated as per the method of Maniatis and Sambrook et al, 1982. 3µL of bromophenol blue loading dye mixed with 7µL of plasmid DNA sample and electrophoresed at 100 V in 0.8% agarose (Himedia) incorporated with 1.5 µL Ethidium bromide (0.1mg/mL) in 1XTris Acetic EDTA (TAE) buffer to confirm the presence of DNA. Further to the confirmation, Restriction mixture contains 3µL plasmid DNA, 3µL EcoRI (Bangalore Genei), 4 µL 10X restriction enzyme buffer and 10 µL double distilled water was prepared in a eppendorf and incubated for 37ºC for 3 hrs. 4 µL of gel loading dye was mixed to the reaction mixture and incubated and cooled at 0ºC to arrest the restriction digestion. Then the samples were loaded in 1.2% agarose with 1.5 µL Ethidium bromide and electrophoresed at 50 V in 1X TAE buffer for 1 hour. The band of the digested DNA was observed by illumination of the gel in UV transilluminator that causes ethidium bromide stained fragments to florescence. Band pattern was interpretated as if there is no digestion single DNA band was seen and in case of extra bands the digestion is incomplete.
Results
The present study revealed that restriction enzyme EcoRI digested the plasmid DNA of E. coli RY13. EcoRI cleaved plasmid DNA in to fragments similar to discrete size and the indication of precise band pattern is the positive result of restriction Fig.1. The effective digestion proved the presence of EcoRI target sequence site in plasmid DNA of E.coli RY13 Fig. 2. EcoRI catalyses and recognizes the six cutters or palindromic sequences of the plasmid DNA Fig. 3.
 |
Figure 1: Agarose gel electrophoresis shows DNA fragment (Lane 1 – 5) of E.coli RY13 Plasmid DNA digested by the restriction enzyme EcoRI. |
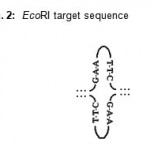 |
Figure 2: EcoRI target sequence.
|
 |
Figure 3: Six cutters or Palindromic sequence
|
Discussion
The purpose of the study is to support the evidenced research of restriction enzyme EcoRI digests plasmid DNA of a significant genetic strain E.coli RY13 by targeting their specific sequence GAATTC. E.coli plasmid DNA is considered as an efficient cloning vector in genetic engineering. Even many restriction enzymes used in recombinant DNA studies were isolated from genetically important strains of E.coli. EcoRI is also isolated and purified from the same strain E. coli RY13. Greene et al. developed a method for the purification of EcoRI endonuclease from E. coli RY13 strain with the yield of 13 U/g cell. Modrich et al has modified this method to increase the yield of the enzyme to 190 U/g cell from the same strain.
In our study, the efficient digestion may be due to the reason that the restriction enzyme EcoRI and plasmid DNA were isolated from the same origin E. coli RY13 strain.
Restriction digestion or mapping is the process of obtaining structural information on a piece of DNA by the use of restriction enzymes. Perhaps, revealing of this structural information in a DNA by restriction mapping helps in the sophisticated research of medical genetics and disease diagnostics.
References
- Helinski DR. Bacterial Plasmids: Autonomous replication and vehicles for gene cloning. CRC Critical rev Biochem. 7, 83 -101 (1979).
- Bolivar F. and Backman K. Plasmids of Escherichia coli as cloning vectors. Methods in enzymol. 68: 245(1979).
- Arber W. and Linn S. DNA modification and restriction. Ann Rev Biochem. 38, 467 – 500 (1969).
- Meselson M. and Yuan R. DNA restriction enzyme from coli. Nature. 217, 1110 – 1114 (1968).
- Tamerler C, Onsan Z, Kirdar B. A Comparative Study on the Recovery of EcoRI Endonuclease from Two Different Genetically Modified Strains of Escherichia coli. Turk J Chem. 25, 63 –71 (2001).
- Yuan R. Structure and mechanism of multifunctional restriction endonucleases. Ann Rev Biochem. 50, 285 – 315 (1981).
- Cheng S, Kim R, King K, Kim S, Modrich P. Isolation of Gram Quantities of EcoRI Restriction and Modification Enzymes from an Overproducing Strain. J Biol Chem. 259, 11571 – 11575 (1984).
- Modrich P and Zabel D. EcoRI Endonuclease: Physical and Catalytic Properties of the Homogeneous Enzyme. J Biol Chem. 251, 5866 – 5874 (1976).
- Greene PJ, Heyneker HL, Bolivar F, Rodriguez RL, Betlach MC, Covarrubias A, Backman LK, Russel DJ, Tait R, Boyer HW. A general method for the purification of Restriction Enzymes, Nucl Acid Res. 5(7), 2371 – 2380 (1978).
- Rubin RA and Modrich P. Purification and Properties of EcoRI Endonuclease, Methods in Enzymol. 65, 96 – 110 (1980).
- Mehra RS, Malhatra VP, Rembhatkar GW. Rapid Purification of a Restriction Endonuclease from Escherichia coli Biotechnology Techniques. 7(6), 411– 414 (1993).

This work is licensed under a Creative Commons Attribution 4.0 International License.





