How to Cite | Publication History | PlumX Article Matrix
V. Shanmugaraju1*, N. Abirami2, A. Mahalakshmipriya1 and K. Selvam1
1Department of Boitechnology, Dr.NGP Arts and Science College, Coimbatore - 641 048 India. 2Department of Boichemistry, Dr.NGP Arts and Science College, Coimbatore - 641 035 India. Corresponding Author E-mail: rajugenes@yahoo.co.in
ABSTRACT: Five marine isolates were isolated from marine water sample of seacoast Kollam Kerala ,India. The isolates were screened for antibacterial activity against Methicillin Resistant Staphylococcus aureus (MRSA) which is a major nasocomical pathogen.The results reveals that among five isolates one isolate(M5)showed the zone of inhibition of 24mm against MRSA .The cell free supernatant of M5 also showed the same activity and the proteinaceous nature of the compound for inhibition of MRSA growth was confirmed by enzyme treatment analysis and further 85KD of protein responsible for the observed activity was visualized while resolving the supernatant under SDS-PAGE. The isolate M5 was identified by 16srDNA sequencing and it reveals that isolate is Lysinibacillus sphaericus The 16srDNA sequence was aligned and phylogenetic relationship was studied.
KEYWORDS: Lysinibacillus sphaericus; antimicrobial peptides; MRSA; antibacterial protein
Download this article as:| Copy the following to cite this article: Shanmugaraju V, Abirami N, Mahalakshmipriya A, Selvam K. Antibacterial Protien (85kda) Produced by Lysinibacillus Sphaericus Against Methicillin Resistant Staphylococcus Aureus. Biosci Biotechnol Res Asia 2009;6(1) |
| Copy the following to cite this URL: Shanmugaraju V, Abirami N, Mahalakshmipriya A, Selvam K. Antibacterial Protien (85kda) Produced by Lysinibacillus Sphaericus Against Methicillin Resistant Staphylococcus Aureus. Biosci Biotechnol Res Asia 2009;6(1) Available from: https://www.biotech-asia.org/?p=7991 |
Introduction
Methicillin Resistant Staphylococcus aureus(MRSA) is the most problematic gram positive bacterium in public health not only because it is highly prevalent but also it has become resistant to almost all available antibiotics except Vancomycin and Teicoplanin[1].Recently, its susceptibility to Vancomycin has decreased and Vancomycin intermediate and Vancomycin resistant Staphylococcus aureus have increasingly been found in clinical isolations in developing and developed countries[2,3,4] Further now a days MRSA isolates exhibiting Teicoplanin resistance has also been reported in several cases .The reports of MRSA resistance to Vancomycin and Teicoplanin,which are antibiotics of last choice makes the need for the suitable alternative antibiotics and drug after Vancomycin treatment have failed during emergency conditions.By viewing the significance of the MRSA resistance against Vancomycin and Teicoplanin. We conducted a screening among marine organisms for anti-MRSA substance producing marine bacteria. The marine environment is a prolific resource for the isolation of less exploited microorganisms. In recent years marine microorganisms have become important in the study of novel microbial products exhibiting antimicrobial ,antiviral, antitumour as well as anticoagulant properties. The active compounds may serve as model systems in the discovery of new drugs.
By considering the scope of marine bacteria and the less exploited nature of marine microorganisms ,in this present study a preliminary screening was made to isolate and identify the bacteria for anti-MRSA substance production and partial characterization of the anti-MRSA substance was also carried out.
Materials and Methods
Sample Collection
The water sample was collected from Kollam, seacoast, Kerala, India The sample was collected in sterilized polythene bags and were brought to laboratory by preventing any contamination on the way .The sample was then stored at a temperature of 4οC until used to minimize the metabolic activities of the microorganisms and to keep them in their exact qualitative and quantitative level of population.
Isolation of Bacteria
The marine water sample was serially diluted by using tenfold dilution .After serial dilution from each dilution 0.1ml of sample was taken and spread plated on marine agar(Himedia,Mumbai).The inoculated plates were incubated at 34ο C for 24 hours to obtain colonies. After the incubation period the individual colonies were picked upon the basis of their macroscopic characters such as size ,shape ,surface appearance, texture and colour. And further the isolates were purified by repeated streaking and were sub cultured on to nutrient agar slants. Then the marine nutrient isolates were stored at 4ο C for further analysis and were designated as M1,M2,M3,M4,and M5.
Screening For Antibacterial Activity Of Marine Isolates By Point Inoculation Method
MRSA was obtained from KMCH clinical laboratory Coimbatore, Tamilnadu. India. The MRSA was checked for its sensitivity pattern by disc diffusion technique against Vancomycin and Methicillin .The overnight grown culture of the MRSA in nutrient broth was uniformly swabbed on the surface of the Muller Hinton Agar (Himedia) plates using sterile cotton swab .Then the marine isolates (M1,M2,M3,M4,M5) were spotted on to the Muller Hinton Agar plates which is seeded with MRSA .The plates were incubated at 34οC for 24 hours. After the incubation period the plate was observed for the zone of inhibition.
Screening of Supernatant for Antibacterial Activity by Agar well Diffusion Method
The marine isolates were grown in marine broth for overnight and cell free supernatant was collected by centrifuging the broth culture at 6000g for 15 minutes .Five wells of 6mm size were made with sterile cork borer on the surface of MRSA seeded Muller Hinton Agar plates.100microliter of the cell free supernatant was added to each well aseptically .The plate was incubated without inverting at 34οC for 24 hours.
Enzymatic Digestion To Confirm Proteinaceous Nature Of Antibacterial Compound
The cell free supernatant was exposed to three different enzymes(Pepsin 10mg/ml,Proteinase K 10 mg/ml and Lipase 20 mg/ml) for overnight to analysis the type of compound causing bacterial growth inhibition .Aliquots(250microliter) of the supernatant were mixed with 250microliter of the enzymes and the pH of the mixture and the incubation temperature was adjusted to get optimal enzyme activity . After 24 hours the pH of all samples were readjusted to 6 to attain maximum antibacterial activity . Well diffusion assay was used to check the antibacterial activity of enzyme supernatant mixture.
Protein Visualization
The homogeneity and molecular weight of protein in denaturing conditions were carried out by SDS-PAGE on 4-15% Polyacrylamide gel electrophoresis as described by Laemmli(1970)[5]. 200microliter of cell free supernatant was loaded along with the protein marker. After electrophoresis proteins were visualized by silver staining.
Isolate Identification
The isolate exhibiting activity against MRSA was identified by 16srDNA sequencing.16srDNA was amplified by polymerase chain reaction using universal primer and sequenced. The PCR was performed with universal 16srDNA primers(5’-AGAGTTTGATCCATGGC- 3’) and (5’-GTTACCTTGTTACGACTT-3’).The used PCR temperature profile was 94ο C for 3 minutes followed by 30 cycles of 98 ο C for 25 seconds,50οC for 1 minute, 72ο for 90 seconds and final extension step at 72ο for 5 minutes. Sequencing of amplified product was carried out in GENEI , Banglore, INDIA. The sequence homology of 16srDNA gene was performed by using Kimura-2 parmeter
Results and Discussion
From water sample five different marine isolates were obtained and the isolates were designated as M1,M2,M3,M4 and M5.All the five isolates were screened for antibacterial activity against MRSA. Among five isolates only one isolate (M5) showed the activity against MRSA. It has produced the zone of inhibition of 24mm(fig1).The cell free supernatant of M5 isolate also showed the same activity against MRSA(fig2) .The enzyme treatment analysis of supernatant proves that the proteinaceous nature of the anti MRSA substance(table1). Inhibition assays revealed that activity was completely lost in the presence of proteinase k and pepsin. This indicates the proteinaceous nature of the compound. Then the protein present in the supernatant was resolved in SDS-PAGE shows the presence of 84KD protein(fig3).16srDNA sequence(fig4) reveals that the isolate M5 is Lysinibacillus sphaericus .The nucleotide similiraty and homology of the Lysinibacillus sphaericus was studied.(table 2,table3)
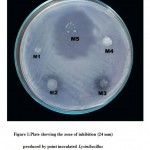 |
Plate 1: showing the zone of inhibition (24 mm) produced by point inoculated Lysinibacillus sphaericus (M5) against MRSA.
|
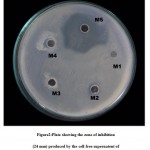 |
Plate 2: showing the zone of inhibition (24 mm) produced by the cell free supernatent of Lysinibacillus sphaericus (M5) against MRSA.
|
 |
Figure 1: Silver stained SDS PAGE showing the 84 kDa antibacterial protien seperated from cell free supernatent of Lysinibacillus sphaericus.Lane A-antibacterial protien and Lane B-Molecular weight marker. |
 |
Figure 2: Aligned 16sr DNA sequence of Lysinibacillus sphaericus(1517 bp).
|
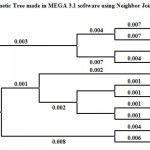 |
Figure 3: Phylogenetic Tree made in MEGA 3.1 software using Neighbor Joining method.
|
Vancomycin resistant MRSA is a major problem in the treatment of nasocomical infection. In the present study a attempt was made to identify a antimicrobial peptide which possess inhibitory activity against such a MRSA .The study report shows a marine isolate has the ability to produce proteinaceous compound responsible for anti MRSA activity .The SDS PAGE analysis reveals that the 85KD protein is responsible for the inhibition of MRSA growth The marine isolate was identified as Lysinibacillus sphaericus by 16srDNA sequencing.
Bacillus amyloliquficians produced antimicrobial peptide subtilosin possess antimicrobial activity against L.monocytogens and S.agalactiae [6] .Recently Shelburne etal(2007)[7]. conducted the study on the spectrum of antimicrobial activity of antimicrobial peptide subtilosin. Their findings suggest that while the bacteriosin has the ability to act on a wide range of organism inefficacy against capsulated organisms limits its practical value. But in the present study the 85KD protein possess the activity against MRSA. Simillarly 85KD protein was isolated by Arlettelongeon[8] which possess activity against Staphylococcus aureus and human pathogenic strains involved in dermatological diseases.James et al [9]identified antibacterial protein from marine bacterium D2which exhibits activity by oligomeric structure and also size exclusion chromatography studies conducted by Arlettelongeon et al reveals 280 kDa of trimer protein which has activity against Staphylococcus aureus .Similarly the 85 kDa protein identified in this study may be assemble either trimer or oligomeric form and inhibit the growth of MRSA .Further studies are required to know the detailed mechanism behind the inhibitory activity of the protein.
The 16srDNA sequencing of isolate confirmed the isolate is Lysinibacillus sphaericus and the sequence was aligned for finding the closest homologous for the microbe. Nearest homologous species was found to be Bacillus sphaericus. The information about the phylogenetic relationship of Lysinibacillus sphaericus with other bacteria were found using combination of NCBI gene bank and RDP databases (fig5)The sequence description reveals that the isolate is phylogenetically related to Bacillus fusiformis strain DSM 2898T,Lysinibacillus sphericus strain C3-41,Bacillus fusiformis strain SW-B9.Bacillus sphaericus strain DSM 28 and Bacillus macroides strain 608.It was found to be more related to Bacillus sphaericus strain DSM 396. Nature has been a source of medical agents, for thousands of years .An impressive number of modern drugs have been isolated from microorganisms in the past century .However an increase in role has been played by microorganisms in the production of antibiotics and other drugs .This finding have important impact for the discovery of novel protein based antimicrobial compounds from marine bacteria and may allow the development of new drug active against MRSA. Further characterization of 85KDa protein is needed to use this in clinical field.
References
- Witte,W;1999;Antibiotic resistance in gram positive bacteria: epidermological aspects. J.Antimicrob.chemother.44:1-9.
- Hanaki,H.,H.Labischinski,Y.Inaba, N.Kondo, H.Murakami,and K.Hiramatsu.1998. Increase in glutamine – non- amidated muropeptides in the peptidoglycan of Vancomycin-resistant staphylococcus aureus strain Mu50. J.Antimicrob.chemother:42:315-320
- Hiramatsu,K,H.Hanaki, Ino, K.Yabuta,T.Oguri,and F.C.Tenover 1997. Methicillin-resistant staphylococcus aureus clinical strain with reduced Vancomycin susceptibility. J.Antimicrob. chemother.40:135-136.
- Patersen, D.L.1999. Reduced susceptibility of staphylococcus aureus to Vancomycin-a review of current knowledge.Commun.Dis.Intell.24:69-73
- Laemmli,U.K(1970) Clevage of structural proteins during the assembly of the head of bacteriaphage T4.Nature 227:680-685.
- Sutyak .K.E., Wirawan.R.E, Aroutcheva.A.A and Chikindas.M.L., 2008; Isolation of Bacillus subtilis antimicrobial peptide subtilosin from the dairy product – derived Bacillus amyloquefaciens; Journal of Applied Microbiology;104(4):1067-1074
- Shelburne CE,AnFY,Dholpe V, Ramamoorthy A, Lopatin DE,Lantz MS. The spectrum of antimicrobial activity of the bacteriocin subtilosin A. J.Antimicrob chemother 2007;59:297-300.
- Arlette Longeon, Jean peduzzi, Michel Barthelemy,Sophie Corre, Jean-Louis Nicolas and Michele Guyot;2004;Purification and partial identification of Novel Antimicrobial protein from marine bacterium pseudoalteromonas species strain X153; Marine Biotechnology; 6(6); 633-641
- James,S.G.,Holmstrom,. and Kjelleberg, S.(1996) Purification and charecterization of novel antibacterial protein from marine bacterium D2.Appl Environ Microbial 62:2783-2788.

This work is licensed under a Creative Commons Attribution 4.0 International License.





