How to Cite | Publication History | PlumX Article Matrix
Insilico and Invitro Studies of Abrin-A Ribosome Inactivating Protein on Candida Albicans
Suma Bhaskar1,2 and T. R. S. Chouhan3
1Dr. M.G.R. Educational and Research Institute (Deemed University), Chennai - 600 095 India.
2Department of Physiology, Adichunchanagiri Institute of Medical Sciences, B.G. Nagara - 571 448, Nagamangala Taluk, Mandya District, Karnataka India.
3Ethicamatrix CRO Pvt. Ltd, Erragadda, Hyderabad - 500 038 India.
Corresponding Author E-mail: suma.bhaskar@gmail.com
ABSTRACT: Increased incidence of fungal infections, particularly those caused by yeast species, has resulted in the development and introduction of a number of new antifungal agents. With the growing problem of antifungal drug resistance, there is a need for the development of effective antifungal agents. With easy access to high speed computers and graphic programs, Computer-assisted drug design (CADD) or molecular modeling has become an essential tool in the process of drug designing. In our present study, we have tried to correlate the results of a molecular docking program Hex 5.1 with that of invitro results. Using Hex 5.1 we have docked Abrin holotoxin, a ribosome inactivating protein with galactose, a compound found aplenty in the cell wall matrix of Candida albicans. Since the results showed the formation of receptor-ligand complex with minimal energy (E= -671.72) and least steric hindrances (-1), indicating favorable interactions, we decided to check whether abrin holotoxin would bring about inhibition of growth of C.albicans invitro by disc diffusion assay. The results of invitro experiments however showed no inhibition of growth of C.albicans in culture in any given dilutions of Abrin holotoxin, inferring that favorable insilico results need not always translate into invitro results.
KEYWORDS: Abrin; R.I.P; RCSB PDB; Hex 5.1; docking; Candida albicans; antifungal
Download this article as:| Copy the following to cite this article: Bhaskar S, Chouhan T. R. S. Insilico and Invitro Studies of Abrin-A Ribosome Inactivating Protein on Candida Albicans. Biosci Biotechnol Res Asia 2009;6(1) |
| Copy the following to cite this URL: Bhaskar S, Chouhan T. R. S. Insilico and Invitro Studies of Abrin-A Ribosome Inactivating Protein on Candida Albicans. Biosci Biotechnol Res Asia 2009;6(1). Available from: https://www.biotech-asia.org/?p=8166 |
Introduction
In the last few decades the incidence of fungal infections by Candida albicans and other related human opportunistic yeast species has increased dramatically due to the rise in the number of immuno-compromised patients (1). Majority of the nosocomial fungal infections are due to Candida species, with C.albicans being the most frequently isolated species. Although it most frequently infects the skin and mucosae, Candida can cause pneumonia, septicemia or endocarditis in the immuno-compromised patients (2). Thus an effective therapeutic intervention is the need of the day. The knowledge that ribosome inactivating proteins bring about apoptosis and that they are internalized through galactose binding sites on the cell wall, the fact that Abrin holotoxin could not attack C.albicans indicate possible deactivation process in C.albicans that needs to be studied to find a possible means of cure to Abrin toxicity. The presence of a large number of mannose and galactose in the form of glycoproteins, glycolipids, and other polysaccharides on the cell wall of C.albicans gave rise to the possibility of using abrin, a powerful plant toxin, as a fungicide. Abrin is a type-II ribosome inactivating protein [RIP] found in the seeds of Abrus precatorius. It is a heterodimer made up of two chains- A and B linked by means of a disulphide bond (Fig. 1). The B chain binds to the cell surface receptors made of glycoproteins and glycolipids containing terminal galactose on the eukaryotic cell surface and facilitates the entry of toxic A-chain into the cell by receptor-mediated endocytosis. The A-chain inhibits protein synthesis through its N-Glycosidase activity by depurinating a specific adenine residue from the GAGA/sarcin/ricin loop of 28S rRNA from 60S ribosome. This region is crucial for ribosomal function, as it is involved in elongation factor-1 (EF-1)-dependent binding of aminoacyl-tRNA and EF-2 catalyzed GTP hydrolysis and translocation. Action of abrin A-chain in the sarcin/ricin loop leads to loss of stability of the loop and thus impairs protein synthesis (3, 4). Besides the inhibition of protein synthesis, abrin has been shown to induce Apoptosis or Programmed cell death (PCD). It brings about PCD by activating Caspase 3; a Caspase known to degrade many structural and regulating proteins as well as proteins involved in DNA repair and also triggers DNA fragmentation leading to cell death by 12 hours. Hence, molecules like abrin and other similar proteins may have tremendous potential as possible therapeutic agents. . Molecular modeling, also known by various names such as Computer-assisted drug design (CADD), Computer-assisted molecular design (CAMD) etc., has become a valuable and essential tool to medicinal chemists in the drug design process. It describes the generation, manipulation or representation of 3-Dimensional structures of molecules and associated physico-chemical properties (5).
In our present study, galactose, which is found in large numbers as galactans along with other polysaccharides on the cell wall matrix of Candida albicans, was chosen to be the receptor. Using a molecular docking program Hex 5.1, Abrin holotoxin (ligand) was docked onto galactose residues on the cell wall of C.albicans. Having obtained promising results for the binding and steric interactions between abrin holotoxin and galactose residue (least energy value, E= 671.72), the computational simulations of ligand-receptor interaction was further evaluated invitro.
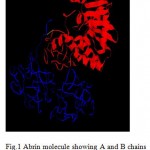 |
Figure 1: Abrin molecule showing A and B chains linked by a disulphide bond (in green).
|
Materials and Methods
The structure of abrin holotoxin and galactose were retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) Protein data bank (PDB) [6]. RCSB PDB is a worldwide repository of information about the 3D structural data, atomic coordinates and other information describing proteins and other important biological macromolecules. Docking is the process by which two molecules fit together in 3-dimensional space. Docking allows one to virtually screen a database of compounds and predict the strongest binders based on various scoring functions. During the course of the process, the ligand and the protein adjust their conformation to achieve an overall “best-fit” like a 3-dimensional hand-in-glove analogy, and this kind of conformational adjustments resulting in the overall binding is referred to as “induced-fit”. Hence docking plays an important role in the “Rational Design of Drugs (RDD)” (7, 8, 9). The docking analysis of Abrin holotoxin with galactose was carried by HEX docking software version 5.1 and their relative stabilities were evaluated using molecular dynamics and their binding affinities, using free energy simulations.
Invitro experiments involved the purification of abrin holotoxin from Abrus precatorius seeds and assessment of putative antifungal activity of abrin holotoxin on the growth of C.albicans was tested by standard disc diffusion assay. Abrin holotoxin was purified from the seeds of Abrus precatorius as described by Lin et al., (10). Fungal growth from the nail bed scrapings from patients with onychomycosis, a chronic, usually painless infection that results in thickening and discoloration of the nail was collected in sterile containers. These were then added to BHI (brain heart infusion) broth and kept for incubation at 37°C for 4 hours. Turbidity was adjusted to 0.5 McFarland. The colony of C.albicans was then uniformly streaked for lawn culture onto Sabouraud dextrose agar plates. 5mm sterile filter paper discs made from Whatman No.3 were then carefully transferred onto the surface of seeded agar plate. 20 µl dilutions of Abrin holotoxin (9.52, 4.76, 2.38, 1.19, 0.595 µg/ml) were pipetted onto the filter paper discs. The plates were then incubated at 37°C for 48 hours following which the diameter of the inhibition zone was measured. The net zone of inhibition was determined by subtracting the disc diameter (i.e. 5 mm) from the total zone of inhibition shown by the test disc in terms of clear halo fungal lawn around the disc. All the experiments were done in triplicates to ensure reproducible results.
Results and Discussions
The docking results tabulated in the Table-1 show that the interactions of abrin holotoxin with the galactose moiety have a high negative energy value. This infers that Abrin holotoxin can be used as the lead molecule since it has maximum interaction having high negative e-value. The table also shows that the bumps are -1, which indicates that the steric clashes are minimal, and therefore it is a favorable interaction.
Table 1: Docking results of Abrin holotoxin with galactose.
| RIP | Receptor | Energy values | Bumps |
| Abrin | Galactose | -671.72 | -1 |
Invitro experiments involving disc diffusion assay showed no inhibition of growth of C.albicans at any given dilution of Abrin holotoxin (Fig. 5), indicating that Abrin holotoxin failed to function as an antifungal agent although theoretical and insilico computational modeling studies show abrin-galactose interactions to be a favorable one.
 |
Figure 2: Harmonic surface of Abrin molecule.
|
 |
Figure 3: Harmonic surface of Galactose.
|
 |
Figure 4: Docking of Abrin with Galactose .
|
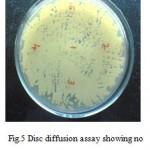 |
Figure 5: Disc diffusion assay showing no inhibition of growth of C.albicans.
|
Conclusion
Molecular docking and invitro results in the present study reveal that the abrin-galactose complex show maximum intermolecular interactions, minimum energy and steric hindrances, and yet, Abrin holotoxin is unable to bring about inhibition of growth of C.albicans in culture. It can be said that although computational docking simulations programs such as Hex provide a useful alternative method to study the protein-protein or ligand-receptor interactions insilico, it is important to emphasize that they cannot substitute for a clear understanding of the system being studied. Such computational simulations can only be considered as an additional tool to gain better insight into the chemistry and biology of the problem at hand.
References
- Fox, J. L. “Fungal infection rates are increasing.” ASM News 59: 515-518 (1993).
- Pfaller, M. A., and Diekema, D. J. “Role of sentinel surveillance of candidemia: trends in species distribution and antifungal susceptibility.” J. Clin. Microbiol. 40: 3551-3557 (2002).
- Endo, Y. and Tsurugi, K. “The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA.” J. Biol. Chem., 263, 8735-8739 (1988).
- Wool, I. G., Gluck, A., and Endo, Y. “Ribotoxin recognition of ribosomal RNA and a proposal for the mechanism of translocation.” Trends Biochem. Sci., 17(7), 266-269 (1992).
- Nadendla, R. R. “Molecular modeling: A powerful tool for drug design and molecular docking.” Resonance. Vol. 9 (5) 51-60 (2004). http://www.pdb.org/
- Kitchen, D. B., Decornez, H., Furr, J. R., Bajorath, J. “Docking and scoring in virtual screening for drug discovery: methods and applications.” Nature reviews. Drug discovery 3 (11): 935–949 (2004).
- Jorgensen, W. L. “Rusting of the lock and key model for protein-ligand binding.” Science 254 (5034): 954–955 (1991).
- Wei, B. Q., Weaver, L. H., Ferrari, A. M., Matthews, B. W., Shoichet, B. K. “Testing a flexible-receptor docking algorithm in a model binding site.” J. Mol. Biol. 337 (5): 1161–1182 (2004).
- Lin, J. Y., Lee, T. C., Tung, T. C. “Isolation of Anti tumor proteins Abrin-A & Abrin-B from Abrus precatorius.” Int. J. Pept. Protein Res. Vol. 12 (5), 311-317 (1978).

This work is licensed under a Creative Commons Attribution 4.0 International License.





