How to Cite | Publication History | PlumX Article Matrix
Cytogenetic Diversity Analysis of Coix Species Using ISSR Markers
A. S. Naik*1 and S. D. Taware2
1Department of Botany, S.B.E.S.College of Science, Aurangabad India. 2Genome Life Sciences, Labh Chambers, Station Road, Aurangabad India. Corresponding Author E-mail: anjali.naik360@gmail.com
ABSTRACT: The genus Coix, a wild relative of maize, belonging to tribe Maydeae of family Poaceae is represented by 3 species in India viz. C. aquatica, C. gigantea and C. lacryma-jobi. It is of wide occurrence in most of south-east Asian countries and of great economic importance as food, forage and medicinal plant. Even though the floras written on Grasses of India describe this genus to be comprised of 3 species, there is not only negligible morphological distinction among these, but at least 2 of them produce a range of semi fertile interspecific hybrids and all form various number of aneuploids. Some interspecific hybrids are suspected to have originated through apomixis. Formation of interspecific hybrids is rather contradictory to the very definition of a species Thus species delimitation poses a taxonomic problem for this genus. The single sequence repeats (SSR) micro satellites, present in nuclear and organelles DNA can be used as molecular markers and has wide ranging applications in the field of genetics including kinship and population studies. These Inter simple sequence repeats (ISSR) represent genome region between micro satellite loci. Sequences amplified by ISSR-PCR can be used for delimiting species.
KEYWORDS: Coix; Cytogenetic diversity; ISSR; phylogeny analysis
Download this article as:| Copy the following to cite this article: Naik. A. S, Taware. S. D. Cytogenetic Diversity Analysis of Coix Species Using ISSR Markers. Biosci Biotechnol Res Asia 2009;6(2) |
| Copy the following to cite this URL: Naik. A. S, Taware. S. D. Cytogenetic Diversity Analysis of Coix Species Using ISSR Markers. Biosci Biotechnol Res Asia 2009;6(2). Available from: https://www.biotech-asia.org/?p=8815. |
Introduction
Coix is an oriental wild relative of maize. It belongs to tribe Maydeae of family Poaceae.It is widely distributed all over South-East Asian countries. In India also, its occurrence has been reported from almost all states, showing its adaptation to a variety of climatic and edaphic conditions.Coix has been extensively studied and exploited from food, fodder and medicine point of view, in countries like China, Japan, Thailand, Indonesia, Korea and Brazil etc. In India, its commercial value as a food crop has been totally neglected, though it has been studied here widely for its cytology and genetics.
The genus Coix is represented by three species viz. Coix aquatica with 2n = 10 chromosomes, C. gigantea with 2n = 20 chromosomes and C. lacryma-jobi with 2n = 20 chromosomes.(Bor N.L.,1960) The species C. aquatica and C. gigantea were found to breed freely and spontaneously, in nature (Deshpande, 1986) and also when controlled cross pollinations were carried out, producing a range of semi-fertile interspecific hybrids in a range from 2n = 11 to 2n = 26 (Naik, 1991). In addition to these, all three species have been reported to produce number of fertile aneuploids (Barve; 1983). Some aneuploids of C. gigantea and C. lacryma-jobi were shown to have been established and successfully competed with their diploid counterparts. (Barve S.S.,1983)
However, there are no clear cut morphological markers on the basis of which the species can be identified in the field. One has to rely upon the conventional cytological method such as study of meiotic prophase-I, especially diakinesis to ascertain their distinction as different species. When the meiotic behaviour of the chromosomes of interspecific hybrids between C.aquatica and C.gigantea was studied, the 5 fairly large and nearly metacentric chromosomes of C.aquatica formed heteromorphic pairs with the chromosomes of C.gigantea genome comprising of two large, four medium sized and four small chromosomes. From a total of four smaller chromosomes of C.gigantea, it is difficult to say which of them are homologous they are of same size and shape. Some chromosomes were left as univalents. The fact that these species hybridize naturally is indicative of their close relationship. The specific delimitation of genus Coix has always posed a problem to taxonomists. All the vegetative characters such as annual vs. perennial habit, leaf breadth, glands on leaf surfaces etc., together with features of reproductive organs like margin of lower glumes of male spikelets, size and shape of the involucres of grain, seem to overlap and appear to be continuous in different species. This has caused much uncertainty with respect to diagnostic features for specific delimitation. It seems that conventional cytological methods should be supplemented with molecular data regarding the DNA analysis to confirm the taxonomic position of Coix species. Through this present investigation, an attempt has been made to explore, whether any molecular markers can be identified which might help in species delimitation in the genus Coix.
Traditionally, classification of the various subgenera, species, and subspecies is based primarily on morphological attributes. However, these traits may not be significantly distinct and usually require growing plants to maturity prior to identification. Moreover, morphological characters may be unstable due to environmental influences. Over the years, the methods for detecting and assessing genetic diversity have extended from analysis of discrete morphological traits to biochemical and molecular traits. Several DNA marker systems are now common use in diversity studies of plants. The most commonly used marker systems are restriction fragment length polymorphism (RFLP) (Soller and Beckmann, 1983), random amplified polymorphic DNA (RAPD) (Williams et al. 1990), amplified fragment length polymorphism (AFLP) (Vos et al. 1995), inter simple sequence repeats (ISSRs) (Zietkiewicz et al. 1994) and microsatellites or simple sequence repeats (SSRs). Among them to characterize DNA variation patterns within species and among closely related taxa in Vigna species have been RAPD (Ba et al. 2004; Dikshit et al. 2007), AFLP (Sivaprakash et al. 2004; Fang et al. 2007), RFLP (Kaga et al. 2000), ISSR (Ajibade et al. 2000), SSRs and sequence tagged microsatellite site (STMS) (Phansak et al. 2005). Inter SSR (ISSR) fingerprinting was developed such that no sequence knowledge was required. Primers based on a repeat sequence and the resultant PCR reaction amplifies the sequence between two SSRs, yielding a multilocus marker system useful for fingerprinting, diversity analysis and genome mapping. PCR products are radiolabelled or PCR incorporation, and separated on a polyacrylamide sequencing gel prior to autoradiographic visualization. concluded that ISSR would be a better tool than RAPD for phylogenetic studies. Nagaoka and Ogihara (1997) have also reported that the ISSR primers produced several times more information than RAPD markers in wheat.
Material and Methods
Karyotypic Analysis
Chromosome Preparation
The seed material of C. auatica, C. gigantea and C. lacryma-jobi was collected from Junnar in Pune district, Ambe ghat near Kolhapur district and Shivaji university campus, Kolhapur, and Thailand respectively. The seeds were sown in large earthen pots on the terrace, in the month of June. The immature male racemes were fixed in Carnoy’s fluid (1: 3 acetic alcohol) for 24 hours and then transferred in straw-yellow colored Ferric chloride solution for mordanting. The anthers were squashed in 1 % acetocarmine to make cytological preparations for the study of meiosis, especially, to confirm their chromosome number.
DNA Amplification condition and gel electrophoresis.
DNA Isolation
Leaf samples of all the three species and four samples were used for DNA isolation and genetic diversity. Genomic DNA was extracted from 4 individual Coix samples according to standard protocol. Isolation of genomic DNA from plant leaf samples was carried out using CTAB method (Doyle and Doyle, 1987)
DNA Quantification:
Quantitative analysis of isolated DNA was carried out using standard Spectrophotometric method for nucleic acid quantification. It was done by Spectrophotometric Assay. The ratio of OD260/OD280 was determined to assess the purity of the sample.
iii. PCR
PCR technique has promoted the development of a range of molecular assay systems which detect polymorphism at molecular level. In this study we used the most widely adopted PCR based ISSR marker technology for characterizing the amongst the Coix sample.
The PCR amplification was carried out in 200 µl reaction mixture tube containing 10X PCR buffer (2.0 µl), 20 ng of genomic DNA (3 µl), 2 µl of UBC ISSR primer (3 pmol/µl), 10 mM dNTPs (2.0 µl), 3U/µl Taq DNA polymerase (0.4 µl), Tween 20 (0.2 µl). The final volume was made to 20 µl by adding SMQ. PCR reactions were carried out in a Finnzyme make thermal cycler.
PCR reactions were carried out in a Finnzyme make thermal cycler using ISSR primers with di and tri nucleotides. ISSR primer sets were ordered from University of British Columbia (UBC). Fourteen ISSR primers were used for initial screening. Out of fourteen primers, 05 primers gave the amplification (in terms of repeatability, scorability) were selected for identification. The selected 4 primers were 14-23 mers. UBC Primer number 813, 814, 815, 819 and 821 showed the genetic variability.
PCR amplified product separated on 1% agarose gel. Gel stained using ethidium bromide and visualized under the UV Transilluminator. For each sample, each fragment / band that was amplified using ISSR primers was treated as a unit rearrangement in genome. The primers which were given scorable and consistently reproducible amplicons were considered. The gel pictures were taken and documented to computer by using Alpha Imager gel documentation system and size of each amplicon was measured by using Alpha Imager Software with respect to standard molecular weight DNA ladder and molecular weight of each of the potential specific bands was calculated using the software program Alpha Imager.
Data Analysis
Marker index for ISSR markers was calculated in order to characterize the capacity of each primer to detect polymorphic loci among the genotypes. It is the sum total of the polymorphism information content (PIC) values of all the markers produced by a particular primer.
The data obtained by scoring the ISSR profiles with different primers individually as well as collectively were subjected to the construction of similarity matrix using Neighbor-joining tree construction method of Nei and Li/Dice. The similarity values were used for cluster analysis. Data analysis was done using Free Tree and Tree View bioinformatics software.
Result and Discussion
Genetic diversity analysis at cytological level
Total of 70 seeds were sown. The results of germination and chromosome number analysis based on conventional cytological methods were as per the following table:
| No. of seeds
Sown |
No. of seeds
Germinated |
Plants obtained, with chromosome numbers:
2n=10 2n=18 2n=20 |
||
| 80 | 76 | 03 | 43 | 30 |
Sample-1
Out of total 70 seeds sown, 3 plants showed chromosome no. 2n=10 with 5 bivalents at diakinesis (Fig.1). All chromosomes are metacentric and fairly large. The rest of meiotic behavior of the chromosomes was normal. This chromosome number was denoted as sample-1
 |
Figure 1: Sample 1, Coix aquatica 2n=10.
|
Sample-2
Out of total 62 seeds that germinated, 26 plants showed chromosome number 2n=18 (Fig.2). These belonged to Coix gigantea .These seeds were collected from Junnar(western Maharashtra).Initially, the populations of Coix gigantea, collected from Taluka Purander, of Dist. Pune and from nearby places of it showed all plants with chromosome nos.2n=20. The plants had showed habit. These 20 chromosomes formed 10 perfectly paired bivalents. When the seeds of these plants collected and raised in the botanical garden of Dr.B.A.M.University, Aurangabad, around the year 1975, gradually number of aneuploids were reported from the natural populations of Coix gigantea.First cytological evidence of non-disjunction of a bivalent (Sapre and Barve, 1985) was reported. This non-disjunction in 2n=20, resulted in producing plants of C. gigantea with chromosome no.2n=18, 2n=19 and, 2n=21 (nullisomic, monosomic and trisomic, respectively). In subsequent years some more aneuploids with chromosome numbers ranging from 2n=18 to 2n=26 (Sapre and Barve, 1983) were also reported. The nullisomics with 2n=18, were not only found to be fertile but also to establish themselves to such an extent that they have completely replaced the 2n=20 (normal diploid chromosome no.) from the population.
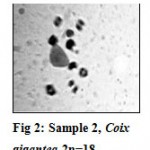 |
Figure 2: Sample 2, Coix gigantea 2n=18.
|
In the present investigation also, seeds collected and plants raised from wild population of C. gigantea, surprisingly showed 2n=18 chromosome number in 43 plants, out of total 46 plants, indicating that, in wild populations also, nullisomics have replaced the diploids with 2n=20 chromosome number. The genome of 2n=18 C.gigantea comprises of 1 pair, 4 medium sized and 4 small sized pairs of chromosomes. One pair of large sized chromosomes got eliminated as a result of meiotic non-disjunction. These plants with chromosome no. 2n=18 were denoted as sample no.2
Sample-3
Some plants of the C.gigantea population, on cytological screening, showed 2n=20 chromosome number, with 10 bivalnts.These were denoted as sample no.3
Sample-4
Seeds were also acquired, by post, from Thailand. On cytological screening of the population raised from these seeds, all 30 plants showed chromosome no. 2n=20 with most of the pollen mother cells showing 10 bivalents and in few of them, 8 II + 1IV (Fig.3). These plants were denoted as Sample-4.
 |
Figure 3: Sample 4, Coix lacryma-jobi 2n=20.
|
SSR markers diversity among Coix
Amplification of genomic DNA of 04 samples using ISSR marker analysis, yielded total amplified 24 fragments. ISSR analysis yielded 24 fragments that could be scored, out of which 01 was polymorphic while the remaining were 23 were monomorphic in nature.
In ISSR analysis number of amplified fragments ranged from 4 to 8 and which varied in size from 255 bp to 1500 bp. UBC primer No. 814 showed the lowest bands while the primer number 821 produced the highest molecular weight bands. Out of the 24 amplified bands produced, only 01 band was polymorphic in nature with 4.16% polymorphism. An average of 1.04% polymorphic fragments per primer was observed. Percentage of polymorphism ranged from 0% (with UBC primer 814, 815 and 821) to 14.28% (with UBC primer 819) (Fig. 4). The PCR amplification using ISSR primers gave rise to reproducible amplification products. The complete data was based on a total of 24 amplicons produced. A Dendrogram analysis was carried out by using bioinformatics phylogeny tool Free Tree and Tree View of DNA fingerprint analysis (Fig. 5). Distance similarity matrix was calculated by using Neighbour-joining tree construction method of Nei and Li/Dice which ranged from 0.0000 to 0.01587 (Tab.1).
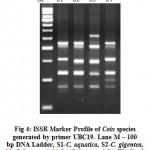 |
Figure 4: ISSR Marker Profile of Coix species generated by primer UBC19. Lane M – 100 bp DNA Ladder, S1-C. aquatica, S2-C. gigentea, S3-C. lacryma-jobi, S4-C. lacryma-jobi (Thailand).
|
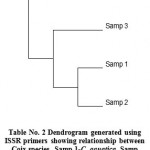 |
Figure 5: Dendrogram generated using ISSR primers showing relationship between Coix species. Samp 1-C. aquatica, Samp 2-C. gigentea, Samp 3-C. lacryma-jobi, Samp 4-C. lacryma-jobi (Thailand).
|
Table No. 1 Distance Similarity Matrix:
Neighbor-joining tree construction method of Nei and Li/Dice.
| C. aquatica | C. gigentea
(2n=18) |
C. gigantea,
( 2n=20) |
C. lacryma-jobi (Thailand) | |
| C. aquatica | 0.00000 | 0.01587 | 0.00000 | |
| C.gigentea,2n=18 | 0.00000 | 0.01587 | 0.00000 | |
| C.gigantea,2n=20 | 0.01587 | 0.01587 | 0.01587 | |
| C. lacryma-jobi (Thailand) | 0.00000 | 0.00000 | 0.01587 |
Discussion
The present investigations showed that during the cytological studies of the Sample 1 of Coix aquatica, out of total 70 plants, 3 plants showed chromosome number. 2n=10 with 5 bivalents at diakinesis. In Sample 2, the wild population of C. gigantea, surprisingly showed 2n=18 chromosome number while few nullisomics have replaced the diploids with 2n=20 chromosome number. The investigations of Sample 3, C. gigantea population, on cytological screening, showed 2n=20 chromosome number, with 10 bivalents. While Sample 4 C. gigantea collected from Thailand showed chromosome number 2n=20 with most of the pollen mother cells showing 10 bivalents and in few of them, 8 II + 1IV. This cytological analysis shows that C. lacryma-jobi there is instability in the chromosome number of the Coix and this varies from 2n=10 to 2n=20. Also the nullisomic condition was observed.
The molecular investigations couldn’t show any such variation at molecular level with the primers used. The bands produced by UBC primer number 814, 815 and 821 does not showed any confirmatory polymorphism. While only primer number 819 has given the polymorphic band. The percentage of polymorphism was not found to be satisfactory. According to the banding pattern with the primers used, the Dendrogram produced showed the evolutionary order of all the four samples.
Acknowledgements
Authors acknowledge to the University Grant Commission (UGC) for the grant of minor research project and the financial support.
References
- Ajibade, S.R., Weeden N.F. and Michite, S. Inter simple sequence repeat analysis of genetic relationships in the genus Vigna. Euphytica, 111:1 (2000), 47-55.
- Ba, F., Pasquet, R.S. and Gspts, P. Genetic diversity in cowpea [Vigna unguiculata (L.) Walp.] as revealed by RAPD markers. Genetic Resources and Crop Evolution, 51:5, (2004),539-550.
- Barve, S.S. Cytogenetical Studies in The Tribe Maydeae, Ph.D. thesis, Dr.B.A.M.University (1983).
- Bor, N.L. The Grasses of Burma, Ceylon, India and Pakistan (1960).
- Deshpande, D.S. Cytology of the interspecific and intergeneric hybrids in the Tribe Maydeae, Ph.D. Thesis, Dr.B.A.M.University, (1986)
- Dikshit, H.K.; Jhang, T.; Singh, N.K.; Koundal, K.R.; Bansal, K.C.; Chandra, N.; Tickoo, J.L. and Sharma, T.R. Genetic differentiation of Vigna species by RAPD, URP and SSR markers. Biologia Plantarum, 51:3, (2007), 451-457.
- Doyle, J.J., and J.L. Doyle.. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19 (1987), 11-15.
- Fang, J., Chao, C.T.; Roberts, P.A. and Ehlers, J.D. Genetic diversity of cowpea [Vigna unguiculata (L.) Walp.] in four West African and USA breeding programs as determined by AFLP analysis. Genetic Resources and Crop Evolution, 54:6 (2007), 1119-1209.
- Kaga, A.; Yoon, M.S.; Tomooka, N. and Vaughan D.A. Collection of Vigna spp. and other legumes from the islands of southern Okinawa prefecture, Japan. In: Report to IPGRI and East Asia Plant Genetic Resources Coordinators. National Institute of Agrobiological Resources, Japan, (2000), 2-25.
- Nagaoka, T. and Ogihara, Y. Applicability of inter simple sequence repeat polymorphisms in wheat for use as DNA markers in comparison to RFLP and RAPD markers. Theoretical and Applied Genetics, 94:5, (1997), 597-602.
- Naik, A.S., Cytogenetical Studies in the Tribe Maydeae, Ph.D.Thesis Dr.B.A.M.University, (1991).
- Phasank, P.; Taylor, P.W.J. and Mongkolporn, O. Genetic diversity in yardlong bean (Vigna unguiculata ssp. sesquipedalis) and related Vigna species using sequence tagged microsatellite site analysis. Scientia Horticulturae, 106:2 (2005), 137-146.
- Sapre, A.B. and Barve,S.S Occurrence of pentasomic and hexasomic plants in wild population of Coix gigantea; Current Science, 52:1; (1983), 614-615.
- Sapre, A.B and Barve, S.S.; Typical meiotic nondisjunction and its consequences in Coix gigantea. The Journal of Heredity; 76 (1985), 387-389.
- Sivprakash, K.R.; Prashanth, S.R.; Mohanty, B.P. and Parida, A. Genetic diversity of black gram landraces as evaluated by AFLP markers. Current Science, , 86:10 (2004), 1411-1415.
- Soller, M. and Beckman, J.S. Genetic polymorphism in varietal identification and genetic improvement. Theoretical and Applied Genetics, 67:1 (1983), 25-33.
- Vos, P.R.; Hogers, R.; Bleeker, M.; Reijans, M.; Lee, T. Van De; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M. and Zabeau, M. AFLP: a new technique for fingerprinting. Nucleic Acids Research, 23:21(1995), 4407-4414.
- Williams, J.G.K.; Kubelik, A.R.; Livak, K.J.; Rafalski, A.J.A and Tingey, S.V. DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research, 18:22 (1990), 6531-6535.
- Zietkiewicz, E.; Rafalski A. and Labuda, D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics, 20:2 (1994), 176-183.

This work is licensed under a Creative Commons Attribution 4.0 International License.





