How to Cite | Publication History | PlumX Article Matrix
Sanjeev Kumar Shukla1*, Shahaj Uddin Ahmed2*, Ashutosh Tiwari2*, Jose Mathew1 and Deepak Sharma2*
1Genome Mapping laboratory, Central Avian Research Institute, Izatnagar, Bareilly India.
2Department of Biotechnology, Bundelkhand University, Jhansi, India.
Corresponding Author E-mail: sanjeevcloning@gmail.com
ABSTRACT: Major Histocompatibility Complex (MHC) class I molecules play an essential role in the immune defense against intracellular infections. This 92-kilobase region of the B locus contains only 19 genes, making the chicken MHC roughly 20-fold smaller than the human MHC. Virtually all the genes have counterparts in the human MHC, defining a minimal essential set of MHC genes conserved over 200 million years of divergence between birds and mammals. The small size and simplicity of the chicken MHC allows co-evolution of genes as haplotypes over considerable periods of time, and makes it possible to study the striking MHC-determined pathogen-specific disease resistance 8-10 at the molecular level. The MHC class I gene was amplified, cloned and sequenced in Red Jungle Fowl (RJF) using primers specific to BF2 gene in chicken. The amplified RJF MHC class I amino acid sequence was 179 amino acids in size. 88 Amino acids in a1 domin (complete exon-2), 91 amino acids in a2 domin (complete exon-3). The Percentage of polymorphism in amino acid sequence of a1 domain in RJF and other poultry species in chicken, guinea fowl, quail, duck and goose was 6.82, 35.56, 30.00, 48.86 and 46.59%, a2 domin 8.79, 17.39, 25.27, 34.78 and 33.70%.
KEYWORDS: CDNA, Red Jungle Fowl (RJF); MHC Class I
Download this article as:| Copy the following to cite this article: Shukla. S. K, Ahmed. S. U, Tiwari. A, Mathew. J, Sharma. D. Molecular Cloning of Major Histocompatibility Complex Class I Cdna From Red Jungle Fowl (Gallus Gallus). Biosci Biotechnol Res Asia 2009;6(2) |
| Copy the following to cite this URL: Shukla. S. K, Ahmed. S. U, Tiwari. A, Mathew. J, Sharma. D. Molecular Cloning of Major Histocompatibility Complex Class I Cdna From Red Jungle Fowl (Gallus Gallus). Biosci Biotechnol Res Asia 2009;6(2). Available from: https://www.biotech-asia.org/?p=8727. |
Introduction
The MHC comprises a group of highly polymorphic genes with a central role in the immune system whose major function is the binding and presentation of foreign antigens to T lymphocytes. The MHC of many species, including birds, is comprised of large multi-gene families as a result of widespread gene duplication1, 2. The B-F/B-L region in the B locus of the chicken MHC is characterized by strong effects on graft rejection, mixed lymphocyte reaction and graft-versus-host reaction and MHC class I genes were found in the Rfp-y region3, 4, 5 nearby on the same microchromosome6. Moreover, the biggest surprise in chicken came from reports that a single dominantly expressed class I locus (BF-IV) determines the immune response to certain infectious pathogens7, 8, 9, 10. Therefore, it is considered to constitute the chicken pendant to the mammalian MHC. The chicken MHC class I molecules are biochemically and structurally similar to the mammalian class I molecules.11, 12
MHC class I molecules are expressed on the surface of virtually all cells of the body and have highly polymorphic class I heavy chain (α chain) and a non-covalently associated non-polymorphic light chain (β2-microglobuin or β2m). MHC is the cluster of gene and is roughly 20-fold smaller than the human MHC, HLA.13 In chicken genes encoding class I molecules are present in MHC proper or B locus and in non-MHC region, known as Rfp-Y or Y locus. The MHC is encoded by number of multigene families. Chicken MHC genes are arranged into two genetically independent clusters, the B system and Rfp-Y. One of the clusters, the B system, was defined initially as a blood group system.14 ,15
In B locus, two class I molecules genes i.e. BF1 and BF2 are present, among which the BF2 is predominantly expressed in chicken. The BF2 gene has been well characterized in chicken. The BF2 gene has been characterized in different B haplotypes in White Leghorn, in commercial broilers16 and in native chicken lines.17, 18 Among other economically important poultry species, MHC class I genes are very well studied in quail, where multiple class I loci i.e. Coja – A, -B, -C and -D were reported19, 20 cloned the MHC class I gene in duck and classified the Anpl-MHC I family genes into four lineages (Anpl-UAA, -0UBA, -UCA and –UDA). Later also characterized MHC class I genes in goose (Ancy-MHC I). Based on genetic distance21, they grouped the Ancy MHC I genes from six individual into four lineges (Ancy-NA, -NB, – NC and –ND). In all these species, the MHC class I genes retained characteristic features of functional MHC class I antigen presentation molecules along with high polymorphism in the amino acid residues in the peptide binding reasons. MHC contain two class I and three class II genes which had lower expression and seemed to be less polymorphic than the genes in the MHC B system cluster. However, little information on the MHC of other birds has been reported.22
Chickens were domesticated from RJF approximately 5,000~7,000 years ago. Chicken and quail would have diverged 36 million years ago23 and their genetically distance was greater than that between chicken and jungle fowls.24, 25 The RJF, formally known as Gallus gallus, is one of four species in the genus Gallus. It is the wild ancestor of the domestic chicken. The reports on MHC class I genes are lacking in this poultry species. Hence the primary aim of our work is to characterize the MHC class I genes with regards to their structural homologies with MHC class I genes of other poultry species including chicken.
Material and Method
Experimental birds
RJF maintained at the Central Avian Research Institute in Izzatnagar, were utilized for cloning and sequencing.
Used GenBank Accession Nos.
In this study, the sequences of BF2 gene from different chicken B haplotype used GenBank Accession Nos. (Table:-2)
Amplification, Cloning and sequencing
The cells used for total RNA isolation were monocytes. The monocytes were separated from the blood using LSM (MP Biomedicals, LLC, Eschwege, Germany) and cultured in RPMI-1640 Medium (JRH, Biosciences, Kansa, USA) supplemented with Fetal Bovine Serum (FBS) and the cells were stimulated with CON A (mitogen) for 1 hour at 37°C in CO2 incubator (5%). The cells were then harvested and the total RNA was isolated using ‘RNAgentsTM – Total RNA isolation system’ (Promega, Madison, WI, USA) and was reverse transcribed using the ‘RevertAidTM – first strand cDNA synthesis kit’ (MBI Fermentas, Hanover, MD, USA).
Primer Designing
The set of primers were designed, forward primer from 5‘ UTR region (SetI FTGGGTGCGGCGGACTTGA), while the reverse primer (SetI-R GCCTCCTTCCCCCACACTCG) was taken from the 5’ end of exon-4 (Fig-1). In the first set, the forward primer was expected to amplify a 673 bp fragment consisting of complete exon-1, exon-2, exon-3 and partial exon-4.
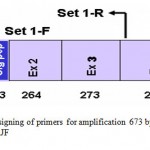 |
Figure 1: Designing of primers for amplification 673 bp of BF2 gene in RJF.
|
 |
Figure 2: Alignment of the translated amino acid sequences of the α1 domains from RJF and other chicken B haplotypes.
|
. “.” same amino acid as in Red Jungle Fowl, = : beta strand; + : alpha helix; P : presumed contacts with peptides; A : presumed contacts with α2 domain; B : contacts with β microglobulin, T : presumed contacts with T cell receptor; # : conserved key amino acids which interacts with antigenic peptide terminal in HLA-A2, respectively; $ : conserved cysteins for disulphide bonds, * : conserved N-glycosylation site. (Fig-2)
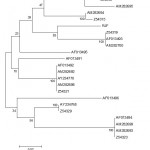 |
Figure 3: Phylogenetic tree based on amino acid variation in α1 and α2 domain between RJF and chicken B haplotypes. Values at nodes represent bootstrap replication scores (based on 100 resamplings).
|
The PCR was performed in a total volume of 25 μl containing 2 μl cDNA, 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl, pH 8.8, 0.1% Triton X-100, 0.01% gelatin, 200 μM of each dNTP, 1 unit of Taq DNA polymerase enzyme (Promega) and 10 Pico mole of each forward and reverse primer. Amplification conditions were 94ºC for 3 min and 35 cycles of 45s at 94ºC, 45s at 56ºC and 1 min at 72ºC and final extension of 10 min at 72ºC. The PCR products were analyzed on 1.6 % agarose gel followed by ethidium bromide staining and visualized under ultraviolet light.
The PCR products were purified from gel using QIAquick Gel Extraction Kit (QIAGEN Inc. Valencia, CA, USA). The purified PCR products were cloned into the pTZ57R/T vector (MBI Fermentas). The positive clones were identified initially by colony PCR and subsequently by insert release after EcoR I and Pst I double-digestion of plasmid DNA. Two representative clones from 4 birds were sequenced on both strands by M13 forward and reverse primers on automated ABI PRISM 3100 advant genetic analyzer (Applied Biosystem, Foster City, CA, USA).
Amino acid sequence analysis
The sequences obtained were first checked manually and blasted (www.ncbi.nlm.nih.gov/BLAST) to ascertain that sequences were of BF2 gene. The related sequences identified from blast results were retrieved from Genbank (www.ncbi.nlm.nih.gov). These sequences were edited and the concerned region i.e. α1 and α2 were cut and saved. Subsequently, the sequences from were α1 and α2 domains were aligned using CLUSTALW website (http://www.cbi.ac.uk/clustalw/). The Molecular Evolutionary Genetic Analysis (MEGA Version 2.1) software was used to estimate nucleotide as well as amino acid variability. The genetic distances between the nucleotide sequences from different poultry species were estimated as Kimura 2- parameter distances, while the genetic distances between the amino acid sequences from different poultry species were estimated as poisson correction distances using MEGA software. Phylogenetic trees were constructed with Neighbour Joining (NJ) procedure using MEGA Version 2.1. Support of the clusters was evaluated by bootstrap, as percentage recurrence of clusters based on 100 bootstrapped replications with MEGA Version 2.1.
Results and Discussion
The interaction of the α1 and α2 domains is formed a groove, known as peptide binding region (PBR). All MHC class I sequences resembled alleles of classical MHC class I genes in having the conserved anchor residues for peptide terminal main chain atoms and amino acid polymorphisms located in the α1 and α2 domains responsible for peptide binding.
The RJF MHC class I amino acid sequence was 179 in size. 88 amino acids in α1 domin (complete exon-2), 91 amino acids in α2 domin (complete exon-3). In chicken, duck and goose 88 amino acids in α1 domin26 however, in quail, it had 90 aa16. While in chicken and quail, it was 91 amino acids in size, duck and goose had 92 aa in α2 domain . Differential selection between genes from different MHC classes could have important implications for disease resistance, as genes of the class I and II regions have different roles in the vertebrate immune response (for instance, the recognition of intracellular and extracellular pathogens, respectively). Many evolutionary studies of MHC variation have relied on the use of diversity measures pooled across several genes27, 28, 29,30. In chicken, the MHC class I genes are very well studied and number of alleles are reported at BF1 as well as BF2 loci in a different chicken lines including White Leghorn, broiler and native chicken. In other poultry species also, while reported four lineages (Anpl-UAA, UBA, – UCA and – UDA)17 of Anpl-MHC I family genes in ducks, also grouped the Ancy MHC class I genes from six individual into four lineges (Ancy-NA, -NB, – NC and –ND)18. The Percentage of polymorphism in amino acid sequence of α1 domain in RJF and other poultry species in chicken, guinea fowl, quail, duck and goose was 6.82, 35.56, 30.00, 48.86 and 46.59%, α2 domin 8.79, 17.39, 25.27, 34.78 and 33.70% .
Within Chicken B haplotype of RJF α1 and α2 domains showed the overall conservation of structure of the PBR region. Two disulphide binding cysteines i.e. C99 and C161 in α2 domain and a potential N-glycosylation site i.e. N85 in α1 domain were conserved. All the conserved residues interacting with the amino terminus of the bonds peptide in the HLA/H2 PBS i.e. Y7, Y59, Y159 and Y171 were conserved in RJF also i.e. Y7, Y58, Y156 and Y168. Three of the four conserved residues, which interacts with the carboxyl terminus of the peptides in HLA/H2 i.e. T143, K146 and W146 were also conserved in RJF i.e. T140, K143 and W144. However, the residue Y84 was not conserved and replaced by R83 in RJF and chicken.
The evolutionary importance of the BF2 locus with respect to disease resistance is supported by evidence that levels of gene expression are tenfold higher than those of the BF1 locus31, 32. Amino acid sequence variation in BF2 gene of α1 and α2 domain within chicken B haplotypes as well as between RJF and within chicken B haplotypes. The amino acid sequence of α1 domain, coded by exon-2 was 88 aa in size and was similar in size the chicken α1 domain. The 24 aa out of 88 aa (27.27 %) were found to be polymorphic within chicken as well as between RJF and chicken. The amino acid sequence of α2 domain, coded by exon-3 was 91 aa in size and was similar in size the chicken α2 domain. The 24 aa out of 91 aa (26.37 %) were found to be polymorphic within chicken as well as between RJF and chicken.
Phylogenetic tree analysis
Phylogenetic tree based on amino acid variation in α1 and α2 domain between RJF and chicken B haplotypes using the ClustalW software.33 Based on this alignment, a phylogenetic tree was constructed using the neighbor-joining method.
The genetic distances (Poisson correction) between RJF and different poultry species were estimated using the cumulative amino acid variability in α1 domain and α2 domain. The estimates are presented (Table-1). Between RJF and different poultry species, genetic distances ranged from 0.083 between RJF and chicken to 0.546 between RJF and duck. Among the poultry species, estimates ranged from 0.273 between duck and goose to 0.596 between quail and goose as well as between quail and duck.
Table:-1 Pair wise genetic distance (poisson correction) between RJF and different poultry species based on cumulative amino acid diversity in α1 and α2 domains.
| RJF | AF0134493 | EU430728 | AB005527 | AB115241 | |
| AF0134493-Chk | 0.083 | ||||
| AF0134493-Chk | 0.318 | 0.318 | |||
| AB005527-Ql | 0.326 | 0.350 | 0.358 | ||
| AB115241-Dk | 0.546 | 0.536 | 0.507 | 0.596 | |
| AY387655-Gs | 0.517 | 0.526 | 0.507 | 0.596 | 0.273 |
The Neighbor-Joining method34 was applies to distance matrices. Phylogenetic tree constructed by using pair wise genetic distances based on nucleotide variability as well as on amino acid variability revealed two major clusters, comprising of guinea fowl, quail, chicken and RJF in one, while duck and goose in other. In first cluster, RJF grouped with chicken, also revealed the clustering of duck and goose separately from other poultry species.35 In first cluster, guinea fowl make separate branch, while chicken and quails are clustered together.Reported the separate clustering of MHC class I sequences from quail and chicken, found that duck MHC class I clusters quite distantly from chicken MHC.
Table 2: Details of sequences of BF2 gene from different chicken B haplotypes used in present study.
| S.No. | Accession Number | Size bp | Type | Haplotype | Reference |
| 1 | Z54315 | 537 | m-RNA | B-F-14 Major | Wallny et al. (2006) |
| 2 | Z54319 | 537 | m-RNA | B-F-21 Major | Wallny et al. (2006) |
| 3 | Z54321 | 537 | m-RNA | B-F-2 Major | Wallny et al. (2006) |
| 4 | Z54323 | 537 | m-RNA | B-F-4 Major | Wallny et al. (2006) |
| 5 | Z54326 | 1289 | m-RNA | B-F12-Major | Wallny et al. (2006) |
| 6 | Z54329 | 537 | m-RNA | B-F-12 Major | Wallny et al. (2006) |
| 7 | Z54360 | 1490 | m-RNA | B-F-19 Major | Wallny et al. (2006) |
| 8 | AM282692 | 3070 | DNA | BF2*0201 | Shaw et al. (2007) |
| 9 | AM282693 | 3080 | DNA | BF2*0401 | Shaw et al. (2007) |
| 10 | AM282694 | 3087 | DNA | BF2*1401 | Shaw et al. (2007) |
| 11 | AM282695 | 3100 | DNA | BF2*1501, | Shaw et al. (2007) |
| 12 | AM282696 | 3097 | DNA | BF2*1902 | Shaw et al. (2007) |
| 13 | AM282698 | 3072 | DNA | BF2*0201 | Shaw et al. (2007) |
| 14 | AM282699 | 3080 | DNA | BF2*0401 | Shaw et al. (2007) |
| 15 | AM282700 | 3090 | DNA | BF2*2101 | Shaw et al. (2007) |
| 16 | AY234768 | 1306 | mRNA | BF12*0201 | Hunt and Fulton (1998) |
| 17 | AY234770 | 1262 | mRNA | BF2*0201 | Hunt and Fulton (1998) |
| 18 | AF013491 | 1262 | mRNA | BFIV-B5 | Hunt and Fulton (1998) |
| 19 | AF013492 | 1262 | mRNA | BFIV-B2 | Hunt and Fulton (1998) |
| 20 | AF013493 | 1262 | mRNA | BFIV-B21 | Hunt and Fulton (1998) |
| 21 | AF013494 | 1262 | mRNA | BFIV-13 | Hunt and Fulton (1998) |
| 22 | AF013495 | 1145 | mRNA | BFIV-17 | Hunt and Fulton (1998) |
| 23 | AF013496 | 1112 | mRNA | BFIV-18 | Hunt and Fulton (1998) |
Acknowledgments
Authors are thankful to Department of Biotechnology, Bundelkhand University, Jhansi (U.P.) India for providing financial support and lab facilities for this study.
References
- Edwards S.V. Hess C.M. Gasper J. and Garrigan D., Toward an evolutionary genomics of the avian MHC. Immunol. Rev. 167, 119-132 (1999).
- Zelano B. and Edwards S.V., An MHC component to kin recognition and mate choice in birds : Predictions, progress, and prospects. Am. Nat. 160, S225-S237 (2002).
- Miller M.M. Goto R. Bernot A. Zoorob R. Auffray C. Bumstead N. and Briles W.E., Two MHC class I and two MHC class II genes map to the chicken Rfp-Y system outside the B complex. Proc Natl. Acad. Sci. USA 91: 4397-4401 (1994)
- Afanassieff M. Goto R.M. Ha J. Sherman M.A. Zhong L. Auffray C. Coudert F. Zoorob R. and Miller M.M., At least one class I gene in restriction fragment pattern-Y (Rfp-Y), the second MHC gene cluster in the chicken, is transcribed, polymorphic, and shows divergent specialization in antigen binding region. J. Immunol. 166, 3324-3333 (2001).
- Rogers S. Shaw I. Ross N. Nair V. Rothwell L. Kaufman J. and Kaiser P., Analysis of part of the chicken Rfp-Y region reveals two novel lectin genes, the first complete genomic sequence of a class I chain gene, a truncated class II chain gene, and a large CR1 repeat. Immunogenetics. 55, 100 (2003).
- Miller M.M. Goto R.M. Taylor R.L. Jr. Zoorob R. Auffray C. Briles R.W. Briles W.E. and Bloom S.E., Assignment of Rfp-Y to the chicken major histocompatibility complex/NOR microchromosome and evidence for high-frequency recombination associated with the nucleolar organizer region. Proc. Natl. Acad. Sci. USA 93, 3958-3962 (1996).
- Kaufman J. and Salomonsen J., B-G we know what it is, but what does it do? Immunol. Today. 13, 1-3 (1992).
- Kaufman J. Andersen R. Avila D. Engberg J. Lambris J. Salomonsen J. Welinder K. and Skjodi K., Different features of the MHC class I hetrodimer have evolved at different rates. Chicken BF and beta 2- microglobulin sequences reveal invariant surface residues. J. Immunol. 148, 1532- 1546 (1992).
- Kaufman J. Milne S. Global T.W. Walker B.A. Jacob J.P. Au Fray C. Zoorob R. and Beck S., The chicken B locus is a minimal essential major histocompatibility complex. Nature. 401, 923- 925 (1999).
- Kaufman J., The simple chicken major histocompatibility complex: life and death in the face of pathogens and vaccines. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 355, 1077-1084 (2000).
- Plachy J. Pink J.R. and Hala K., Biology of the chicken MHC (B complex). Crit. Rev. Immunol. 12, 47-79 (1992).
- Fulton J.E. Thacker E.L. Bacon L.D. and Hunt H.D., Functional analysis of avian class I (BFIV) glycoproteins by epitope tagging and mutagenesis in-vitro. Eur. J. Immunol. 25, 2069-76 (1995).
- MHC Sequencing Consortium Complete sequence and gene map of a human major histocompatibility complex. Nature. 401, 921-923 (1999).
- Briles W.E. McGibbon W.H. and Irwin M.R., On multiple alleles affecting cellular antigens in the chicken. Genetics. 35, 633-652 (1950).
- Okada I., The B complex in the chicken -Development from a blood group system into the major histocompatibility complex. J. Fac. App. Bio. Sci.. Hiro.Uni. 31, 11-28 (1992).
- Livant E.J. Brigati J.R. and Ewald S.J., Diversity and locus specificity of chicken MHC B class I sequences. Ani. Gen. 35, 18-27 (2004).
- Yan R.Q. Li X.S. Yang T.Y. and Xia C., Structures and homology modeling of chicken major histocompatibility complex protein class I (BF2 and beta2m). Mol. Immuunology. 43, 1040-46 (2005).
- Lima-Rosa C.A.V. Canal C.W. Streck A.F. Freitas L.B. Delgado-Canedo A. Bonatto S.L. and Salzano et, al., B-F DNA sequence variability in Brazilian (blue egg Caipira) chicken. Ani. Gen. 35, 278-284 (2004).
- Shiina T. Oka A. Imanishi T. Hanzawa K. Gojobori T. Watanabe S. and Inoko H., Multiple class I loci expressed by the quail MHC. Imm. 49 (5), 456-460 (1999).
- Xia C. Lin C.Y. Xu G.X. Hu T.J. and Yang T.Y., cDNA cloning and genomic structure of the duck (Anas platyrhynchos) MHC class I gene. Imm. 56, 304-309 (2004).
- Xia C. Hu T. Yang T. Wang L. Xu G. and Lin C., cDNA cloning, genomic structure and expression analysis of the goose (Anser cygnoides) MHC class I gene. Vet. Imm. Imm. 107 (3-4) : 291-302. (2005).
- Kulski J.K. Shiina T. Anazai T. Kohara S. and Inoko H., Comparative genomic analysis of the MHC : the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol. Rev. 190, 95-122 (2002).
- Tuinen M. and Hedges S.B., Calibration of avian molecular clocks. Mol. Bio. Evol. 18, 206-213 (2001).
- Nishibori M. Hayashi T. Tsudzuki M. Yamamoto Y. and Yasue H., Complete sequence of the Japanese quail (Coturnix japonica) mitochondrial genome and its genetic relationship with related species. Ani. Gen. 32, 180-185 (2001).
- Nishibori M. Hayashi T. Tsudzuki M. Yamamoto Y. and Yasue H., Phylogenetic analysis of the domestication process in chickens based on the polymorphism of the complete mitochondrial genome DNA. DNA poly. 9 : 110-114. (2001).
- Hunt H.D. and Fulton J.E., Analysis of polymorphism in the major expressed class I locus (BFIV) of the chicken. Imm. 47, 456- 467 (1998).
- Kurtz J. Kalbe M. Aeschlimann P.B. Haberli M.A. Wegner K.M. Reusch T.B. and Milinski M., Major histocompatibility complex diversity influences parasite resistance and innate immunity in sticklebacks. Proc. R. Soc. Lond. B. Biol. Sci. 271, 197-204. (2004).
- Hansson B. and Richardson D., Genetic variation in two endangered Acrocephalus species compared to a widespread congener: estimates based on functional and random loci. Anim Conserv 8, 83-90. (2005)
- Richardson D.S. Komdeur J. Burke T. and Schantz T.V., MHC based patterns of social and extra-pair mate choice in the Seychelles warbler. Proc. R. Soc. Lond. B. Biol. Sci. 272, 759-767 (2005).
- Westerdahl H. Waldenstrom J. Hansson B. Hasselquist D. Schantz T.V. and Bensch S., Associations between malaria and MHC genes in a migratory songbird. Proc. R. Soc. Lond. B. Biol. Sci. 272, 1511-1518 (2005).
- Wallny H.J. Avila D. Hunt L.G. Powelld T.J. Patricia R.P. Salomonsen J. Skjodt K. Vainio O. Vilbois F. Wiles M.V. and Kaufman J., Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC detemined response to Rous sarcoma virus in chickens. Proc. Natl. Acad. Sci. USA. 103, 1434-1439 (2006).
- Shaw I. Powell T.J. Marston D.A. Baker K. Hateren A.V. Riegert P. Wiles M.V. Milne S. Stepha B. and Kaufman J., Different evolutionary histories of the two classical class I gene BF1 and BF2 illustrate drift and selection within the stable MHC haplotypes of chickens. J. Immunol. 178,5744-5752 (2007).
- Saitou N. and Nei M., The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406 (1987).
- Thompson J.D. Higgins D.G. and Gibson T.J., CLUSTAL W : improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nuc. Aci. Res. 11, 22 (22) ,4673-80. (1994).
- Singh, S.K., Genetic polymorphism in BLB2 region and its association with immunocompetence and production traits in guinea fowl (Numida meleagris). PhD Thesis submitted to Bun. Uni., Jhansi, India (2009).

This work is licensed under a Creative Commons Attribution 4.0 International License.





