How to Cite | Publication History | PlumX Article Matrix
Anbalagan Ezhilarasi* and Narayanaswamy Anand
Centre for Advanced Studies in Botany, University of Madras, Guindy Campus, Chennai - 600 025 India.
Corresponding Author E-mail: anbuezhilarasi@gmail.com
ABSTRACT: RAPD techniques were used for the detection of genetic heterogeneticity among the axenic culture of fresh water filamentous heterocystous Anabaena spp. Those were morphologically discriminated two groups, each containing five Anabaena species based on the proximity of the akinetes to heterocyst, adjacent to or away from the spore in the trichome. The electrophoretic patterns for 10 Anabaena strains were used for molecular analysis using the RAPD technique. A total of 10 cyanobacterial isolates were selected and mass cultured in BG11 medium. Genomic DNA was extracted from fresh water cyanobacterial isolates and was amplified using primers CRA 22, CRA 23, CRA 25, OPC11, OPA13, and OPA18 and distinct PCR fingerprint were generated. Unique banding patterns were observed from all tested cyanobacterial species and their molecular weights of each band were used to calculate their genetic distance among them. Random amplification of polymorphic DNA (RAPD) was carried out for the phylogenetic characterization of these strains. RAPD fingerprinting results clearly showed the genetic variation among the cyanobacterial isolates.
KEYWORDS: Anabaena; Cyanobacteria; Randomly Amplified Polymorphic DNA (RAPD); Phylogeny; Taxonomy
Download this article as:| Copy the following to cite this article: Ezhilarasi. A, Anand. N.Phylogenetic Analysis of the Genus Anabaena by Single Randomly Amplified Polymorphic DNA (RAPD) Fingerprinting. Biosci Biotechnol Res Asia 2009;6(2) |
| Copy the following to cite this URL: Ezhilarasi. A, Anand. N.Phylogenetic Analysis of the Genus Anabaena by Single Randomly Amplified Polymorphic DNA (RAPD) Fingerprinting. Biosci Biotechnol Res Asia 2009;6(2). Biosci Biotechnol Res Asia 2009;6(2). Available from: https://www.biotech-asia.org/?p=8911. |
Introduction
Cyanobacteria are among the most widespread, morphologically distinct and abundant prokaryotes known. They are oxygenic photosynthetic autotrophs with the unique ability to fix atmospheric nitrogen, (1, 2,3) originally considered as a class of algae, the blue-green algae. These quality help cyanobacteria inhabit almost all possible biotype, ranging from seawater, deserts and hot springs to Antartic lakes (4). They have an extraordinary biosynthetic potential and a repertoire of diverse metabolic activities. They are one of the dominant genera in various ecological habitats, especially in rice fields, where they are found as both free-living and symbiotic with the water fern, Azolla. The genus Anabaena was established by Bory in 1922. Geitler (5) described 57 European species of Anabaena, while Desikachary (6) designated 25 species in Cyanophyta. Morphology, developmental and biochemical parameters may vary with environmental or culture conditions. They can be classified on the basis of morphology, cellular differentiation, biochemical, physiological and genetic criteria.
DNA base composition is a very important genetic character to study the taxonomy of cyanobacteria. Analysis of the DNA base composition (Mol % G+C) is one of the few molecular characters that have been determined for almost 200 cyanobacterial strains (7). Large differences in DNA base composition indicate that the strains cannot be closely related, whereas similar G+C percentages give no clue concerning genotyping relationships (8). Cyanobacterial genera namely Anabaena, Nostoc, Phormidium, Microcystis, Synechococcus and Synechocystis have been analyzed using molecular techniques such as DNA sequencing, random amplified polymorphic DNA (RAPD) and DNA polymorphisms (9, 10, 11, 12, 13). DNA amplification fingerprinting has been very useful for distinguishing closely related genotypes and is a novel method for classification (14, 15). Analysis of DNA typing results obtained by the rapd method clearly distinguishes the genera Anabaena from Microcystis. The choice of primers with high G+C content provides good DNA profiles at a low concentration and RAPD-PCR have been used to generate uniqueness in phylogenetic analysis of toxic cyanobacteria (10).
In a study by Neilan (10) two 10-mer oligonucleotides were used to develop specific and repeatable DNA fingerprints for cyanobacterial isolates. The use of RAPD analysis was done to distinguish genotypes in five species of Microcystis (M. novacekii, M. viridis, M. wesenbergii, M. aeruginosa and M. ichthyoblabe) (16). Genomic fingerprints have been obtained by PCR with random (RAPD) or arbitrary primers. The morphological and genetic variation using RAPD-PCR of Nodularia strains from Australia and from other geographical sites have indicated the existence or genetically distinct geographical strains (17). Grouping based on the sequence identity was supported by morphological features (size and morphology of vegetative cells, heterocyst and akinetes, and diameter and morphology of trichomes) (17, 18). The genetic variability of the cosmopolitan, ubiquitous fresh water cyanobacterium, Phormedium retzii, was assessed using random amplified polymorphic DNA (RAPD) marker and 16S rDNA sequences (12).
In the present study the genus Anabaena Bory was chosen for taxonomic analysis. The morphological criteria traditionally used for identification of Anabaena species are: biometric characters of vegetative cells, heterocysts and spores. An important feature for species identity of the taxa is the proximity of the akinetes to heterocysts (19,20). In the investigation, our objective was to develop an easy and reliable method to analyze the morphological and genetic variations using RAPD-PCR analyses and finally analyze the genetic diversity in this genus.
Materials and Methods
Organisms and culture conditions
Ten cultures of Anabaena were selected from the Culture Collection of Algae, Centre for Advanced Studies in Botany, University of Madras, India. The original habitats, taxonomic details and akinete positions of the strains are listed in Table 1. These cultures were axenized by Imipenem (Merck), a broad spectrum β-lactam antibiotic (21). All the isolates were maintained in BG 11o medium (1) under a light intensity of 40 µEmˉ²s¯¹ and 25 ± 1ºC. The culture rack was fitted with Sangmo Weston S650 313F automatic model timer to provide alternative light and dark phases of 12 h each. Batch cultures of the isolates were maintained in BG 11o medium in 250 ml Erlenmeyer flasks. Sixteen days old cultures were used for the study.
Preparation of DNA sample for electrophoresis
DNA extraction was carried out according to standard procedures (22). Exponentially growing (50 ml) cells were pelletted by centrifugation and resuspended in 0.5 ml of lysis solution (25% sucrose, 50 mM Tris – HCl, 100 mM EDTA). The cells were treated with 5 mg of lysozyme for 30 min at 37°C. Sodium dodecyl sulfate and proteinase K were added to final concentrations of 1% and 100 µg ml-1, respectively and the samples were incubated at 45°C overnight. The DNA was extracted three times with Phenol: Chloroform: Isoamyl alcohol (25:24:1) and twice with Chloroform: Isoamyl alcohol (24:1). The DNA was precipitated, washed with 70% ethanol, resuspended in 100 µL of Tris – EDTA buffer, and stored at -20°C. Polymerase chain reactions (PCRs) were performed on an ERICOMP, Delta cycler I™ system, Easy cycler ™ PCR system. Oligonucleotides were purchased from DDT, UK.
Randomly Amplified Polymorphic DNA (RAPD) – PCR and electrophoretic analysis
The standard, optimized PCR was performed in a total volume of 50 µl containing 3 mM MgCl2, 200 mM each deoxynucleotide triphosphate (dATP, dCTP, dGTP, and dTTP), 10 pmol of each PCR primer, 1U Taq DNA polymerase, and 10 ng of genomic DNA or 2 µL of cyanobacterial cell lysate supernatant. Thermal cycles were as follows: initial denaturation at 94°C for 4 min, 30 cycles of 94°C for 20 s, 45°C for 30 s, 72°C for 60 s, and final extension at 72°C for 5 min. The random 10 mer oligonucleotide (Decamer) primers were used for this study. Primers viz., CRA 22 – CCGCAGCCAA, CRA 23- GCGATCCCCA, CRA 25- AACGCGCAAC, OPA 11- CAATCGCCGT, OPA13- CAGCACCCAC and OPA 18- AGGTGACCGT were obtained from commercial sources (Integrated DNA technologies, Inc, Coralville) by using previously published data (10). PCR products were resolved on 1% agarose gel in Tris-0.5 X TBE (Tris Borate – EDTA). Agarose gels were stained in an ethidium bromide solution (0.5 mg / mL) and were viewed under the illumination of UV light in a Vilber Loumart gel documentation system.
Data Analyses
Fingerprints generated from different Anabaena species were compared and all bands were scored. The presence or absence of particular DNA fragments was converted in to binary data and the Jaccard distance index was used to obtain the similarity matrix. The tree was constructed using the unweighted pair-group method using arithmetic averages(UPGMA) program in phylogeny inference package (PHYLIP) (23).
Results and discussion
RAPD – PCR analysis (single primer reactions)
Genomic DNA of the different Anabaena species was amplified using RAPD primers viz., CRA22, CRA23, CRA25, OPA11, OPA13 and OPA18 (Fig.1). The electrophoretic pattern for 10 strains of cyanobacteria derived from six-single primer reactions were analyzed to provide diagnostic fingerprint for each culture and genetic distances between strains based on RAPD markers. The DNA fingerprints produced consists of bands between 100 bp to 1000 bp and were clearly distinguished among the strains tested. Each band was considered a genetic marker for the strain from which it was amplified. The data produced from 10 strains of Anabaena were used to calculate genetic distances by pairwise comparisons (data not shown) and the phenogram was constructed. The tree illustrates the similarity of RAPD patterns seen on the gel. From ten strains of Anabaena a total 262 RAPD bands were generated using 8 primers. Of the 8 primers selected, six primers generated informative and reproducible bands in all strains while the two primers, CRA26 and OPA08, did not produce any informative RAPD patterns. Morphological variation did not correlate with phylogenetic variation for any of the six primers tested. RAPD profiling of the genomes has been widely accepted as a valid taxonomic and phylogenetic tool (10). Bolch et al., (18) brought out the genotypic distinction between most toxic and non-toxic Nodularia strains using RAPD-PCR and cpcBA-lGS sequence data. Cyanobacterial G+C content is in the range of 40% to 70%, with the average for Microcystis species genomes being 40% to 43% G+C (24). The primers synthesized in this study were chosen because of their previous usefulness when applied to both plant and bacterial genomes (25, 26). In this study, genotypic diversity of Anabaena strains based on RAPD gene revealed that morphological variations may be dissimilar at the genetic level. The RAPD primers generated fingerprint patterns that were unique for the different cyanobacterial cultures with varying degrees of polymorphism. Distinct finger patterns were obtained with the primers CRA22, CRA23, CRA25, OPA11, OPA13 and OPA18. The phenogram clearly supported the delineation of the genera Anabaena, as shown by the primary bifurcation of the tree. Relatively large genetic distances are seen in the RAPD data, among strains of the species Anabaena (Fig. 2). Neilan (10) reported combination of two primers and comparison of single and multiple-primer reactions. In the present study reactions of six single RAPD primers were analyzed and several new products were synthesized. Van Coppenhole et al. (27) reported the use of three primers separately to provide enough polymorphic information to identify species of the symbiotic genus Anabaena and to create a phylogenetic tree with a topology similar to that derived with 22 primers. The technique used in this study was also performed with a non-radioactive method for the RAPD-PCR products or hybridization. Urbach et al. (28) showed that phylogenetically closely related strains of Prochlorococcus could be physiologically distinct. Further studies on the phylogeny of these organisms need to be carried out by analyzing the products of the primers used alone or in various combinations, which may help in understanding the genetic differences in these unique organisms and may also give an insight into the relationship between the morphological and genetic variations obtained.
 |
Figure 1
|
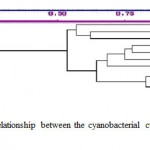 |
Figure 2
|
Table 1: Anabaena strains selected for the study.
|
Taxonomic designation of Anabaena Bory (No. strains) |
Position of heterocysts and akinetes |
Origin / Source
|
|
A.ambigua (A485)
A. torulosa (A525)
A. cylindrica (A621)
A.augstumalis A802)
A. sphaerica (A904)
A. inaequalis (A487)
A. fertilissima (A549)
A. variabilis (A514)
A. subtropica (A618)
A. verrucosa (A622) |
heterocysts adjacent to akinetes
heterocysts adjacent to akinetes
heterocysts adjacent to akinetes
heterocysts adjacent to akinetes
heterocysts adjacent to akinetes
heterocysts away from the akinetes
heterocysts away from the akinetes
heterocysts away from the akinetes
heterocysts away from the akinetes
heterocysts away from the akinetes |
1403/7 CCAP, UK
M2/2 aS2T2 Gif Sur Yvette from, France
Isolate175,Kantz(1403/2a),ICC, U.S.A
Czech.Jahnke 4a
1616 ICC, U.S.A
1403/9 CCAP, UK
M2/3b Gif Sur YvettePCC, France
1403/12 CCAP, UK
Isolate 45 Kantz Feb.71 ICC, U.S.A
ICC, U.S.A
|
CCAP
Culture Collection of Algae and Protozoa,
Cambridge,U.K.
ICC
Indiana Culture Collection, U.S.A.
Acknowledgements
I gratefully acknowledge Ministry of Environment and Forests, Government of India, New Delhi for supporting me with financial assistance through out my research project.
References
- Rippka R, Deruelles J, Waterbury J.B, Herdman M. and Stanier R.Y., Journal of General Microbiology., 111, 1-61(1979).
- Whitton BA., Diversity, ecology and taxonomy of the cyanobacteria. In: N.H. Mann and N.G. Carrs (eds.). Photosynthetic prokaryotes. Plenum (1992).
- Holt J.G, Krieg N.R, Sneath P.H.A, Stanley J.T. and Williams S.T., Bergey’s manual of determinative bacteriology: group 11, oxygenic phototrophic bacteria., 377-475(1994).
- Sigler W.B, Bachofen R. and Zeyer J., Environmental Microbiology., 5, 618-627(2003).
- Geitler L., Cyanophyceae. In: Rabenhorst L (ed.), Kryptogamenflora Von Deutschland, Oesterreich and der Schweiz, vol XIV, Leipzig: Akademische Verlagsgesellschaft (1932).
- Desikachary T.V., Cyanophyta. Indian Council of Agricultural Research, New Delhi, 686 (1959).
- Herdman M, Janvier M, Waterbury J.B, Rippka R, Stanier R.Y. and Mandel M., Journal of General Microbiology., 111, 63-71(1979).
- Wilmotte A., Molecular evolution and taxonomy of the cyanobacteria. In D.A.Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands, 1- 25(1994).
- Mullins T.D, Britschgi T.B, Krest R.L. and Giovannoni S.J., Limnology and Oceanography., 40, 148-158 (1995).
- Neilan B.A., Applied and Environmental Microbiology., 61, 2286-2291(1995).
- Neilan B.A, Jacobs D, del Dot T, Blackall L.L, Hawkins P.R, Cox P.T. and Goodman A.E., International Journal of Systematic Bacteriology., 47, 693-697(1997).
- Casamatta D.A, Vis M.L. and Sheath R.G., Aquatic Botany., 77, 295-309(2003).
- Shalini Dhar D.W. and Gupta R.K., World Journal of Microbiology and Biotechnology., 24, 927-935(2007).
- Welsh J. and McClelland M., Nucleic Acids Research., 18, 7213-7218(1990).
- Williams J.G.K, Kubelik A.R, Livak K.J, Raafalski J.A. and Tingey S.V., Nucleic Acids Research., 18, 6531-6535(1990).
- Nishiara H, Miwa H, Watanabe M, Nagashima M, Yagi O.and Takamura Y., Bioscience, Biotechnology, and Biochemistry., 61, 1067-1072(1997).
- Bolch C.J.S, Blackburn S.L, Neilan B.A. and Grewee P.M., Journal of Phycology., 322, 445- 451(1996).
- Bolch C.J.S, Orr P.T, Jones G.J. and Blackburn S.I., Journal of Phycology., 35, 339-355(1999).
- Anand N., Journal of Madras University., 41, 15-19 (1978).
- Anand N., Acta Botanica Indica., 7, 22-28 ((1979).
- Ferris M.J. and Hirsch C.F., Applied and Environmental Microbiology., 57, 448-1452 (1991).
- Sambrook J, Fritsch E.F. and Maniatis T., Molecular cloning laboratory manual 2nd ed. Cold Spring Harbor Laboratory Press Cold Spring Horbor, N.Y(1989).
- Felsenstein J., PHYLIP (Phylogeny inference Package) version 3.5c.Distributed by the author. Department of Genetics, University of Washington, Seattle, WA, (1993).
- Fahrenkrug P.M, Bett M.B. and Parker D.L., International Journal of Systematic Bacteriology., 42, 182-184(1992).
- Akopyanz N, Bukaanov N.O, Westblom T.U, Kresovicch S. and Berg E., Nucleic Acid Res.,20, 5137- 5142 (1992).
- Kresovich S, Williaams J.G.K, McFerson J.R, Routman E.J. and Schaal B.A., Theoretical and Applied Genetics., 85,190-196(1992).
- Van Coppenhole B, Watanabe I, Van Hove C, Second G, Huang N. and McCouch S.R., Genome., 36, 686-693(1993).
- Urbach E, Scanlan D.J, Distel D.L, Waterbury J.B. and Chisholm S.W., Journal of Molecular Evolution., 16, 188-201(1998).

This work is licensed under a Creative Commons Attribution 4.0 International License.





