How to Cite | Publication History | PlumX Article Matrix
Screening and Isolation of Antibiotic Producing Actinomycetes from Marine Samples Using Rifampicin
Feleke Moges1,2*, T. Prabhakar1, T. Ramana2 and G. Sankar1
1Pharmaceutical Biotechnology Division, A.U. College of Pharmaceutical Sciences, Andhra University India.
2Department of Biotechnology, College of Science and Technology, Andhra University, Visakhapatnam - 530 003 India.
ABSTRACT: The total number of actinomycetes recovered from different samples varied between a maximum of 3.5 X 105 CFU/gm of samples of sea weeds and algae to a minimum of 6.2ª102 CFU/ml of sea water. A total of 132 strains of actinomycetes were isolated, of these 76 (57.6%) exhibit antimicrobial activity to one and/ or more of bacterial, filamentous fungi or yeast strains. Most of the inhibitory activity was directed against bacteria than filamentous fungi and yeast. Among bacterial group Bacillus subtilis (39.4%) was the most susceptible while Proteus vulgaris (11.4%) was the least. Among fungal strains, Penicillium chryseogenum (34.1%) was the most susceptible while Aspergillus oryzae (10.6%) was the least. Thirty five (26.5%) of the total isolates exhibited broad spectrum activity. The result showed that, better recovery of actinomycetes was observed using rifampicin than using naldixic acid. Therefore, in the search for screening and isolation of bioactive producing actinomycetes, rifampicin might be used as a better drug of choice than naldixic acid for suppressing and inhibiting growth of non-filamentous bacterial contaminants in marine samples.
KEYWORDS:
Actinomycetes; marine samples and Rifampicin
Download this article as:| Copy the following to cite this article: Moges. F, Prabhakar. T, Ramana. T, Sankar.G.Screening and Isolation of Antibiotic Producing Actinomycetes from Marine Samples Using Rifampicin. Biosci Biotechnol Res Asia 2009;6(2) |
| Copy the following to cite this URL: Moges. F, Prabhakar. T, Ramana. T, Sankar.G.Screening and Isolation of Antibiotic Producing Actinomycetes from Marine Samples Using Rifampicin. Biosci Biotechnol Res Asia 2009;6(2) . Available from: https://www.biotech-asia.org/?p=8776. |
Introduction
It is well established fact that microbial natural products remain one of the most important sources of lead compounds for the pharmaceutical industry. Among microorganisms, the actinomycetes are a fascinating group that they are sources of most of the antibiotics used in medicine today. They also produce metabolites that are used as anticancer drugs, antihelminthics and drugs that suppress the immune system in patients who have received organ transplants. This becomes apparent in 1940, following Selman Waksman’s discovery of actinomycin from S. gresous1 and was fully realized by the 1980s, when actinomycetes accounted for almost 70% of the world’s naturally occurring antibiotics2.
In addition to terrestrial sources actinomycetes have been isolated from marine water, sediments, plants and animals. Actinomycetes comprise about 10% of bacteria colonizing marine aggregates3. Despite their abundance, however, reports in marine actinomycetes are yet not explored. As a result in recent years marine environment be comes a major focus in the search for the next generation of pharmaceutical agents4. This is because; organisms in marine environment have developed unique adaptations that enable them to survive in dark, cold and highly pressurized environments. These novel adaptations can offer a wealth of opportunities for the discovery of new drugs for the treatment of infectious diseases 5, 6, 7.
The goal of this study were to screen and isolate antibiotic producing actinomycetes that are found in different samples of marine environment in the East Coast of Bay of Bengal using refampicin or naldxic acid as a selective inhibitor of bacterial contaminant.
Materials and Methods
Sample collection and processing
A total of 37 different samples were taken from marine environment (10 sea water, 20 sediments, 1sample each from sponges, hard corals and snails, and 4 algae and sea weeds) at a various depth using sterile screw capped bottle and sterile plastic bags.
Samples were processed as soon as collected and some of the samples collected from distant site were processed in the following days of collection by preserving the samples with a sterile plastic bag in an ice box. Algae and sea weeds samples were dried overnight in a laminar flow hood and crushed with an alcohol sterilized mortar and pestle. Other samples like sponges, corals and shell of snail samples were processed as described by Jensen et. al. 8. These samples were subjected overnight in a laminar flow hood and the samples were scrapped with a sterile spatula generating a powder.
About 1gm of the above samples (sediments, dried powdered samples) was suspended aseptically and were homogenized with sterile sea water and kept in 250 ml flask having 50 ml of sterile sea water and incubated in an orbital shaker at 26 0C with shaking at 140 rpm for 30 minutes. Mixtures were allowed to settle then ten fold serial dilutions were prepared. Depending on the condition water samples were either directly or serially diluted.
One millilitre of each of these dilutions was added to 50ml of starch casein agar (g/l: starch 10, casein 0.3, KNO3 2, NaCl 2, K2HPO4 2, MgSO4.7H2O 0.05, CaCO3 0.02, FeSO4.7H2O 0.01, agar 20) and oat meal agar (g/l: oat meal 20, agar 20, trace salt solution 1ml) when the temperature is about 40-45 0C. Samples were thoroughly mixed and poured in to a sterile Petri-plate (6 diam). One copy of the samples was plated following spread plating technique using sterile L-shaped glass rod. Inoculated samples were incubated at 280C for 21 days. To inhibit growth of non-filamentous bacteria and fungi medium was supplemented with 5µg/ml rifampicin8 or 20µg/ml naldixic acid and 50µg/ml cyclohexamide 9 respectively.
Identification of actinomycetes
Actinomycetes colonies were recognized by their characteristics tough leathery colonies that adhered to the agar surface, branched vegetative mycelia, and when present, aerial mycelia and spore formation 8,10. Actinomycetes colonies were counted on different plates after 10-21 days incubation at 28 0C. Data are expressed as colony forming unit (CFU/ml) for water or CFU/gm for dry weight samples. Actinomycetes colonies were characterized morphologically and physiologically following the methods given in the International Streptomyces Project (ISP) 11. Species were identified by the morphological characteristics of colonies, substrate and aerial mycelium, structure of spore chains and pigment production on different ISP media. Detailed physiological and biochemical characterization of promising isolates were performed following the standard procedure12.
Determination of antimicrobial activity
Primary screening of antimicrobial activities of actinomycetes isolated from different samples were screened following the streak plating technique. The test organisms employed in this study were supplied by the National Chemical Laboratory (NCIM), Pune and Microbial Type Culture Collection and Gene bank (MTCC), Chandigarh. The following test organisms were used for antimicrobial activities. Bacillus pimilus (NCIM-2327), Bacillus subtilis (NCIM-2063), Staphylococcus aureus (NCIM-2079), Escherichia coli (NCIM-2065), Shigella flexinari (MTCC-1457), Salmonella typhimerium (MTCC-98), Aspergillus niger (NCIM-548), Aspergillus oryzae (NCIM-643), Penicillium cryseogenum (NCIM-738), Candidia albicans, and Saccharomyces cervisae.
Further screening of activities were made using an agar overlay methods following methods of Willims et.al. 1983a 13. That is, spot inoculated 5 days old plated colonies were killed by inverting them over 1.5ml chloroform for 40 minutes. The dead colonies were overlaid with 5ml of nutrient agar (bacteria) and 5 ml of potato dextrose agar (yeast and fungi) that has been inoculated with the test organism. Zone of inhibition around the colonies was recorded after 24 hours at 37 0C for bacteria and 48 hours at 28 0C for yeast and filamentous fungi.
Result and Discussion
The total number of actinomycetes recovered from different samples showed that the maximum population density was recorded in samples of sea weeds and algae, 3.5 X 105 CFU/gm followed by samples of sponges, corals and snails, 4.7 X104 CFU/ml; sediments 2.6X103 CFU/ml and water with least population counts, 6.2X102CFU/ml. (Table -1). Sahu et.al14 observed similar patterns of population density of marine actinomycetes from sediment and water samples collected in the East coast of India. The maximum count of actinomycetes in algae and sea weeds samples may be because of nutrient availability. That is, nutrient availability in plant and animal residues is higher and this may create a fertile ground for spores of actinomycetes to germinate. However, the population of actinomycetes decrease in water samples than other samples, this is also because dormancy of spore imposed by lack of nutrients in water and a spore population will decline15 and results low number of actinomycetes in water sample than other samples.
Tables 1: Total microbial count obtained in different samples using Rifampcin.
| Type of sample | Total number of samples | Average total count of actinomycetes | Medium used | Antifungal and antibacterial agents |
| Water | 10 | 6.2X102 CFU/ml
|
Starch casein agar | Cyclohexamide 50µg/ml and rifampicin5µg/ml |
| Sediment | 20 | 2.6X103 CFU/gm
|
Starch casein agar | Cyclohexamide 50µg/ml and rifampicin5µg/ml |
| Sponges, corals and snails | 3 | 4.7X104CFU/gm
|
Starch casein agar | Cyclohexamide 50µg/ml and rifampicin5µg/ml |
| Algae and sea weeds | 4 | 3.5X105 CFU/gm
|
Starch casein agar | Cyclohexamide 50µg/ml and rifampicin5µg/ml |
To avoid bacterial contaminant from different samples, 5μg/ml rifampicin or 20μg/ml naldixic acid have been used for comparison. Comparing to naldixic acid, the rate of recovery of antibiotic producing actinoomycetes using rifampicin is relatively high (Table-2). Similar report by Pisano et.al.16 showed that, in pre-treated samples 16 % of the actinomycetes produced bioactivity while in rifampicin treated marine samples 46% of actinomycetes showed bioactivity17.
As indicated in table-2, four water samples were taken for comparison of rifampicin and naldixic acid. It was found that actinomycetes colonies were recovered only in one sample using rifampicin (1.5X101CFU/ml). Out of 10 sediment samples taken for comparison, actinomycetes were recovered in 5 samples for rifampicin (1.1X103 CFU/gm) and 3 samples for naldixic acid (1.3X102 CFU/gm). One sample each from corals, sponges and shells were also compared, both rifampicin (4.7 X104 CFU/gm) and nalixic acid (2.8X103 CFU/gm) were showing similar recovery with high population density in rifampicin. Four samples from algae and sea weeds were also compared, 3 of the samples were showing high density count using rifampicin (3.6X105 CFU/gm) and 2 of the samples showing also good count using naldixic acid (3.0X104 CFU/gm). As naldixic acid, rifampicin is also effective against some gram-positive and gram-negative bacteria. However, rifampicin is mostly effective when there is combination of other drugs (Examples, treatment of tuberculosis, leprosy); this might be the reason that using rifampicin alone may favour recovery of more actinomycetes populations than using the broad spectrum antibiotic, naldixic acid.
Table 2: Comparison of actinomycetes recovery from same samples using rifampicin and naldixic acid as inhibitors of bacterial contaminants. (Only 21 samples were compared)
| Type of sample | Number of samples assessed | Antibiotics used | Recovery of actinomycetes
|
||
| Yes | No | Average count | |||
| Water | 4 | 5μg/ml Rifampicin | 1 | 3 | 1.5X101CFU/ml |
| 20μg/ml Naldixic acid | –
|
4 | – | ||
| Sediment | 10 | 5μg/ml Rifampicin | 5
|
5 | 1.1X103 CFU/gm |
| 20μg/ml Naldixic acid | 3 | 7
|
1.3X102 CFU/gm
|
||
| Corals, sponges, shells | 3 | 5μg/ml Rifampicin | 2 | 1
|
4.7 X104 CFU/gm |
| 20μg/ml Naldixic acid | 2 | 1 | 2.8X103 CFU/gm | ||
|
Algae and sea weeds |
4 |
5μg/ml Rifampicin |
3 | 1 | 3.6X105 CFU/gm |
|
20μg/ml Naldixic acid |
2 | 2
|
3.0X104 CFU/gm
|
||
During screening of 37 different marine samples, 420 suspected actinomycetes colonies were recovered. Among these, on the basis of their colony morphology, colour of their aerial mycelium 150 colonies suspected to be different were selected and looping out. Although rifampicin can suppress the growth of some of the strains of actinomycetes, 5μg/ml concentration added to the sample during primary screening found to be effective to protect rapidly growing members of non-filamentous bacteria.
After detail study of their morphology and characteristics of aerial and substrate mycelium, a total of 132 isolates were confirmed to be actinomycetes (Table-3). Among these, 76 (57.6%) exhibit antimicrobial activity to one and/ or more test organisms of bacterial, and filamentous fungi or yeast strains while the other 56 (42.4%) isolates don’t show and/or poor activities. This result was lower than reported by Pisano et.al.18, 73% of chitinolytic actinomyctes were showing bioactivity. The reason may be due to correlation between chitinolysis and bioactivity of actinomycetes 18.
Most of the inhibitory activity was directed against bacteria than filamentous fungi and yeast. As shown in table-3, 64 (48.9%) of the total number isolated, proved to be active to one or more bacteria, while 50 (37.9%) were active to filamentous fungi and yeast.
Table 3: Distribution of actinomycets exhibiting antimicrobial activities.
| Type of test organisms | Number of actinomycetes | % |
| Gram positive
Bacteria Gram negative Gram positive and negative Total number of isolates |
27
5 32 64 |
20.5
3.8 24.2 48.9 |
| Filamentous fungi
Fungi Yeast Filamentous fungi & yeast Total number of isolates |
19
2 29 50 |
14.4
1.5 22.0 37.9 |
| Number of isolates with broad spectrum activity (bacteria, filamentous fungi and yeast) |
35 |
26.5 |
| Total actinomycetes showing activities
Total actinomycetes with weak and/or no activities |
76
56 |
57.6
42.4 |
Among bacterial group B. subtilis (39.4%) was the most susceptible organism followed by S. aureus (37.1%), B. pimilus (28.8%), E. coli (25%); P. vulgaris (11.4%) was the least susceptible. Among fungal group Penicillium cryseogenum (34.1%) was the most susceptible followed by Aspergillus niger (18.2%), Saccharomyces cervisae (17.4%), Candidia albicans (14.4%); Aspergillus oryzae (10.6%) was the least susceptible (Figure-1). Total number of actinomycetes exhibiting broad spectrum activity (bacteria and fungi) were 35(26.5%).
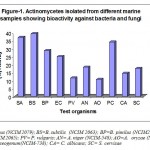 |
Figure 1: Total number of actinomycetes exhibiting broad spectrum activity (bacteria and fungi) were 35(26.5%).
|
SA= S. aureus (NCIM 2079); BS=B. subtilis (NCIM 2063); BP=B. pimilus (NCIM2327); EC= E. coli (NCIM 2065); PV= P. vulgaris; AN= A. niger (NCIM-548); AO=A. oryzae (NCIM 643); PC= P. cryseogenum(NCIM-738); CA= C. albicans; SC= S. cervisae
Fifteen promising isolate were selected and their detailed biochemical and physiological study showed that 10 of them were under the Sterptomyces species and 5 of them were under the group Micromonospora species. This result was in agreement with reports of Das et.al., 19 and Peela et.al., 20 as the major genera isolated in the bay of Bengal were the Streptomyces and Micromonospora species. However, this result is different from reports by Bredholt et.al.21 that Micromonospora species are dominant. The reason may be because of difference in depth of the sample collected. The deep water sediments contained a higher relative amount of Micromonospora compared to the shallow water sediments22. Although, the other isolate showed strong antagonistic activities against different test organisms two of the Streptomyces species were also strongly active against methicillin resistant S. aureus (MRSA (Table-4). The antimicrobial spectrums of these species were also evaluated by submerged fermentation and their activities are still very promising against the test organism of bacterial and fungal strains. The diversity of these promising isolates and their content of biological activities are understudy.
Table 4: Activity of promising isolates on different test organisms using an agar overlay method.
| Code of isolate | Inhibition zone (mm) | ||||||||||||
|
Filamentous fungi and yeast |
Gram-positive and gram-negative bacteria
|
||||||||||||
| AN | AO | PC | SC | CA | SA | BM | BS | EC | PV | SF | ST | MRSA | |
| WS1/31 | 25 | 20 | 28 | 25 | 15 | 15 | 25 | 20 | – | – | – | – | – |
| SS4/2 | – | – | – | – | – | 50 | 20 | 55 | – | – | – | – | – |
| SS4/4 | – | _ | _ | _ | _ | – | 20 | – | – | – | – | – | – |
| SS4/61 | _ | _ | 15 | _ | _ | – | – | 20 | – | – | – | – | – |
| SS16/2 | 15 | – | 35 | 12 | 23 | 32 | 15 | 57 | 45 | 25 | 45 | 47 | 32 |
| SS18/2 | – | – | – | – | – | 10 | 25 | 15 | – | – | – | – | – |
| SS18/3 | – | – | 10 | 30 | 23 | 60 | 25 | 65 | 25 | – | 30 | – | – |
| CS19/10 | – | – | – | – | – | 55 | – | – | 23 | – | 55 | – | – |
| CS19/11 | – | – | – | – | – | 40 | 50 | – | 30 | – | 28 | – | – |
| CS19/13 | – | – | – | – | – | 55 | – | 45 | 23 | – | 28 | – | 30 |
| SS23/2 | 18 | 20 | 40 | 18 | – | 60 | 60 | 70 | – | – | 23 | – | – |
| SS31/1 | – | – | – | – | – | 30 | 45 | 25 | 10 | – | – | – | – |
| SS31/2 | – | – | – | – | – | 48 | 50 | 41 | – | – | – | – | – |
| SS31/32 | – | – | – | – | – | 47 | 45 | 42 | – | 40 | – | – | – |
| SS31/T2 | – | – | – | – | – | 42 | 45 | 38 | – | – | 55 | – | – |
AN= A. niger (NCIM-548); AO=A. oryzae (NCIM 643); PC= P. cryseogenum; SC= S. cervisae; CA= C. albicans; SA= S. aureus (NCIM 2079); BP=B. pimilus (NCIM2327); BS=B. subtilis (NCIM 2063); EC= E. coli (NCIM 2065); PV= P. vulgaris; SF= S. flexinari (MTCC1457); ST=S. typhimerium (MTCC 98); MRSA= Methicillin Resistant Staphylococcus aureus.
Conclusion
As it is well documented drug resistance pathogens such as methicillin resistant S. aureus become the most problematic gram positive bacterium in public health not only because it is highly prevalent but also it has reported that, the organism shows reduced susceptibility to the last resort antibiotics of vancomycin and teicoplanin. Therefore, despite the challenges of identifying new natural product antibiotics; searching and screening of antibiotic producing actinomycetes from new unexplored area using different pre-treatment of marine samples such as rifampicin as selective inhibitors for non filamentous bacterial contaminants during primary screening may help to identify new novel bioactive molecules.
Acknowledgment
The authors are thankful to DBT, New Delhi for the financial support to carry out this work. We are also grateful to Dr. B. Narasinga Rao (King George Hospital) for providing methicillin-resistant S. aureus.
References
- Waksman, S.A. and Woodruff, H.B. Bacteriostatic and bactericidal substances produced by soil actinomycetes. Proc. Soc. Exp. Biol.Med. 45:609-614 (1940).
- Okami,Y. and Hotta K. Search and discovery of new antibiotics, P.33-67. In:M. Goodfellow, S.T.Williams and M. Mordarski (ed), Actinomycetes in Biotechnology, Academic press. Inc., New York (1988).
- Baltz, R.H. Antimicrobials from actinomycetes: Back from the future. Microbe. 2:125-130 (2007).
- Bull A.T., Ward, A.C. and Goodfellow M. Search discovery strategies for biotechnology: The paradigm shift. Micobiol. Mol. Biol. Rev. 64:573-606 (2000).
- Proksch,P., Edrada, R.A., Ebel, R. Drug from the seas-current status and micobiological implications. Applied Micobiol Biotechnol.59:125-134. (2002).
- Newman, D.J. and Cragg, G.M. Marine natural products and related compounds in clinical and advanced preclinical trials. J. Natural products. 67: 1216-1238 (2004).
- Lam, K.S. Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 9:245-251(2006).
- Jensen, P.R, Gontang, E., Mafnas, C., Mincer, T.J., Fenical, W. Culturable marine actinomycets diversity from tropical Pacific Ocean sediments. Environ Microbiol. 7:1039-1048 (2005).
- .Imada C., Noseki N., Kamata M., Kobayashi T. and Hamada-Sato N. Isolation and characterization of antibacterial substances produced by marine actinomycetes in the presence of sea water. Actinomycetologica. 21:27-31. (2007)
- Ghanem, N.M.,Sabry, S.A., El-Sherif, Z.M. and Abu El-Ela, G.A. Isolation and Enumeration of marine actinomycetes from sea water and sediments in Alexanderia. J. Gen. Appl. Micobiol. 46: 105-111 (2000).
- Shirling , E.B., Gottlieb, D. Methods for characterization of streptomyces species. Int. Syst. Bacteriol., 16:313-340 (1966).
- Salle, A.J. “ Laboratory Manual on Fundamental Principles of Bacteriology” 3rd ed. Mc Graw-Hill Book Co. (1948).
- Williams, S.T., Goodfellow, m., Alderson, G.,Wellington, E.M.H., Sneath, P.H.A. and Sackin, M.J. Numerical classification of streptomyces and related genera. J. Gen. Microbiol. 129:1743-1843 (1983a).
- Sahu MK., Kumar KS and Kannan L. Isolation of actinomycetes from various samples of the Vellar Estuary, Southeast Coast of India. Poll Res. 24:45-48 (2005).
- Cross, T. Acquatic Actinomycetes: A critical survey of the occurrence, growth and role of actinomycetes in aquatic habitats. J. Appl. Bacteriol. 50:397-423 (1981).
- Pisano MA, Sommer MJ, Lopez MM. Application of pre-treatments for the isolation of bioactive actinomycetes from marine sediments. Appl Microbiol. Biotechnol. 25:285-288 (1986).
- Pisano MA,Sommer MJ, Brancaccio L. Isolation of bioactive actinomycetes from marine sediments using rifampicin. Appl Microbiol. Biotechnol. 31:609-612 (1989).
- Pisano MA, Sommer MJ. and Taras, L. Bioactivity of chitinolytic actinomycetes of marine origin. Appl Microbiol Biotechnol. 36:553-555 (1992).
- Das S., Lyla P.S. and Ajmalkhan S. Distribution and generic composition of culturable marine actinomycetes from the sediments of Indian continental slope of Bay of Bengal. Chinese J. Oceanology and Limnology. 26:166-177 (2008).
- Peela S. Kurada V. and T. Ramana. Studies on antagonstic marine actinomycetes from the Bay of Bengal. World J. Microbiol. Biotechnol. 21:583-585 (2005).
- Bredholt H., Fjaervik E., Johnsen G. and Zotchev S.B. Actinomycetes from sediments in the Trondheim Fjord Norway: Diversity and biological activity. Mar. Drugs. 6:12-24 (2008).

This work is licensed under a Creative Commons Attribution 4.0 International License.





