How to Cite | Publication History | PlumX Article Matrix
Febriani1, Rukman Hertadi1, Prihardi Kahar2, Akhmaloka1 and Fida Madayanti1*
1Biochemistry Research Group, Faculty of Mathematics and Natural Sciences, Institut Teknologi Bandung, Ganesha 10, Bandung Indonesia.
2Asian Center For Environmental Research, Meisei University, 2-1-1, Hodokubo, Hino-Shi Tokyo, 191-8506 Japan.
Corresponding Author E-mail:fida@chem.itb.ac.id,
ABSTRACT: A local thermophilic microorganism namely DMS-3 has been identified as thermostable alkaline lipase producing isolate. The bacterium was isolated from Domas hot spring, Tangkuban Perahu Mount, West Java. Based on gram staining test and Scanning Electron Microscopy, the microorganism showed a rod shape and a gram positive bacterium. The isolated showed maximum expression of lipase at pH 9 and 70oC. The lipase produced by DMS-3 isolate was purified using column chromatography of DEAE sepharose fast flow and Sephacryl S-200. Following Sephacryl separation the enzyme still showed specific activity at 54 times higher compared to that the wild type, with yield was about 4.89 %. Polyacryamide gel electrophoresis combining with zymogram analysis showed that there were 4 bands of protein exhibiting lipase activity. Further analysis by eluting of each band showed that the first, second, and third band revealed few bands similar of each other on denatured SDS-PAGE. Meanwhile the fourth band only showed single band. The data suggested that DMS-3 isolated expressed more than one type of lipases.
KEYWORDS: Thermostable alkaline lipase; microorganism thermophile; domas isolate.
Download this article as:| Copy the following to cite this article: Febriani, Hertadi R, Kahar P, Akhmaloka, Madayanti F. Isolation and Purification of Novel Thermostable Alkaline Lipase from Local Thermophilic Microorganism. Biosci Biotechnol Res Asia 2010;7(2) |
| Copy the following to cite this URL: Febriani, Hertadi R, Kahar P, Akhmaloka, Madayanti F. Isolation and Purification of Novel Thermostable Alkaline Lipase from Local Thermophilic Microorganism. Biosci Biotechnol Res Asia 2010;(2).Available from: https://www.biotech-asia.org/?p=8971 |
Introduction
Lipases are widely known enzymes, which able to catalyze triglyceride into diacyl glyceride, glycerol, and free fatty acid in hydrolysis process. The enzyme commonly works on interface of water-lipid micellar or emulsion formed by non-polar substrate (long chain triacyl glycerol) and water solvent1-2. It is also recognized that lipases perform reversible reaction so that the enzyme is widely used in hydrolysis process, glyceride trans-esterification, enantioselective synthesis and also hydrolysis of various ester groups3. Within the condition of insufficient water, lipases catalyze the reverse reactions that are esterification, alcoholysis, and acidolysis. Even though lipases have various substrates some lipases shows very specific catalytic properties such as chemo, region, and enantionselective reaction4.
Lipase is one of important enzymes and used in industries, such as detergent, pharmaceutical, food, textile and waste treatment industries5-6. The detergent industry has emerged as one of the major consumers of hydrolytic enzymes especially their work at alkaline pH. At the moment lipases are used more than a quarter of global enzyme production. The use of alkaline lipase is increased remarkably after the availability of lipase source, derived from thermophilic bacteria.7Alkaline lipase is an important enzyme since it has activity at high pH. Now days, alkaline lipases are mostly isolated from Geobacillus sp8-9. The application of lipases in detergent industries require characteristics factors such as stable on high temperature, active in alkaline condition and tolerant to additive materials such as surfactants and denaturation by proteolysis10.
Many novel lipases are still to be explored and identified. The enzymes with new and exciting properties could be discovered from thermophilic microorganisme11. Indonesia is one of the most tectonically active area in the world, has highly potential as habitat of thermophiles12. A few thermophilic microorganisms were isolated from various hot springs around West Java, such as, Domas 13, Papandayan 14 and Kawah Hujan (Kamojang) 12-15 hot springs. Some of these bacteria are identified as thermostable alkaline lipase producer strains16-17.
In this report, we describe the isolation and purification of thermostable alkaline lipase from local isolate of thermophilic bacterium obtained from Domas hot spring in West Java, namely DMS-3. The enzymes have been purified by chromatography of DEAE sepharose fast flow and Sephacryl S-200.
Material and Methods
Material
p-nitropheny palmitate, pepton, yeast extract, NaCl, bacto agar, aceto nitril, etanol ammonium sulfate (NH4)2SO4 were purchases from Sigma. DEAE-Sepharose Fast Flow and Sephacryl S-200 were purchased from Pharmacia.
Morpholological Analysis
A local isolate of thermophilic bacteria which was previously isolated from hot spring in Domas, West Java Indonesia was regenerated on solid medium of ½ T containing 0.8 % polypepton, 0.4 % yeast extract, 0.2 % NaCl, and 3 % bacto agar. The media was then incubated for 18 hours at 70 oC. For scanning electron microscope analysis the samples were fixed for 18 h in 0.25% glutaraldehyde (in Na-phospate pH 7.2). It was subsequently dehydrated in a graded series of ethanol, critical point dried, gold coated by sputtering (ion Sputter CE102 Hitachi) and observed with S-4700 scanning electron microscope. Colony morphology was determined with microscope with 400 x zoom using cultures grown on agar plates for 24 h at 70 °C. Gram staining was performed according to Dussault18.
Isolation of Lipase
A single colony from the solid medium was transferred onto a 25 mL liquid medium contaning 0.5% peptone, 0.5 % yeast extract, 0.05 % NaCL, 0.05 % CaCl2 as starter media. The pH was adjusted up to 9.0 by Glysin-NaOH buffer. The inoculated media was incubated at 70 oC on shaker incubator, 150 rpm for 24 hours. 100 µL of inoculums bacteria was transferred to 100 mL of production starter media. The supernatant of the culture after centrifugation (12000 g, 10 min) at 4 oC was used to determine extracellular lipase activity as crude enzyme.
Enzyme Purification
The cell free supernatant was fractionated by precipitation with ammonium sulfate at 20 – 60% of saturation. All subsequent steps were carried out at 4 °C. The protein pellet was resuspended in 0.02M Tris-HCl buffer, pH 8.0 and dialyzed against the same buffer to remove the residual ammonium sulfate.
The precipitated enzyme was load onto a column of DEAE-sepharose fast flow (2.2 x 20 cm), previously pre-equilibrated with 20 mM Tris-HCl buffer pH 8.0. The column equilibrated with 20 mM Tris-HCl buffer (pH 8.0 0.15 M NaCl). The column was eluted with 20 mM Tris-HCl buffer pH 8.0. It was subsequently eluted with step gradient of 0.15 M-0.8 M NaCl (0.15; 0.3; 0.6; 0.8 M NaCl) in 20 mM Tris-HCl buffer pH 8.0 at flow rate of 2 mL/minute. The fractions exhibiting higher lipase activity were collected.
The pooled enzyme from DEAE sepharose fast flow was concentrated by membrane filters through 10 kDa-cut off (Amicon) and loaded to a Sephacryl S-200 column (1,2 X100 cm), equilibrated with 20 mM tris-HCl buffer (pH 8.0 0.15 M NaCl) at 1 mL/minute. Enzyme activity and protein concentration of pooled fraction was determined. Specific activity of enzyme was calculated in every step of purification.
Native and SDS PAGE
Sample proteins were analyzed on native and sodium dedocyl sulfate-polyacrylamide gel electrophoresis based on Laemmli 19 to check the purity and determine the molecular mass of the purified enzyme. Zymography analysis was carried out for detection of lipase activity. The enzyme was assayed and visualized on zymograms using 12 % Native-polyacrylamide gels. The protein samples containing lipase were washed in 100 mM Tris–HCl buffer pH 7.5 for 15 minute at 4 oC. The gels were finally incubated for 1 hour at 65 oC in developing solution consisting of 3 mM α-naphthyl acetate, 1 mM Fast Red TR (Sigma), and 100 mM sodium phosphate buffer, pH 8.0.
Enzyme Assay
Lipase activity was determined using spectrophotometric assay as described by Lee et al 20 with p-nitropheny palmitate as a substrate. The enzyme activity was measured by monitoring the change in absorbance at 405 nm that represents the amount of released p-nitrophenol (PNP). One unit of lipase is defined as the amounts of enzyme releasing 1 umol PNP per min under the assay condition. Protein concentration was determined by the method of Bradford using Bovine Serum Albumin as the standard 21.
Results and Discussion
Cell Morphology
Morphology of the isolate was analyzed using gram Staining and Electron Microscopy (SEM) methods. Based on the gram staining analysis, the isolate showed as a gram positive bacterium. In addition the isolate show rod shape form with specific clear zone within the cell (Figure 1 A). Furthermore, SEM analysis of the isolate showed that the bacterium appear as long and slim rod shape form (Figure 1B). SEM analysis (Figure 1 B) shows that the cell has uniform. This is suggested that the cell is single strain. The result of morphological analysis is in agreement with the ribotyping analysis result15.
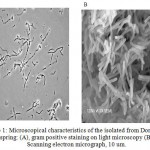 |
Figure 1a: Microscopical characteristics of the isolated from Domas hot spring: (A), gram positive staining on light microscopy (B) Scanning electron micrograph, 10 um.
|
Production of Thermostable Alkaline Lipase
An extracellular alkaline lipase was produced on the media consisting of 0.5% peptone, 0.5 % yeast extract, 0.05 % NaCL, and 0.05 % CaCl2. The lipase was produced optimally at 70oC, pH 9.0 on the late of exponential or the early of stationary phase. 5.115 g of lipase with specific activity of 22.07 U/g are isolated from 1500 mL of culture (Tabel 1). The crude enzyme was then purified.
Purification of Thermostable Alkaline Lipase
Purification of the enzyme was carried out by using ammonium sulfate fractionation followed by DEAE sepharose fast flow and Sephacryl S-200. Ammonium sulfate fractionation was carried out at 0-20, 20-40, 60-80% saturation respectively. Based on fractionation result, we collected ammonium sulfate fraction at 20-60%. Following purification by Sephacryl S-200, the specific activity of purified enzyme still showed 54 times higher compared to that the crude extract, with the yield of 4.9 % (Table 1). The result suggested that have good quality of purified lipase 7-20.
Tabel 1: Summary of thermostable alkaline lipase purification.
| Purification
step |
Sample
volume (mL) |
Protein | Enzyme | |||||
| Concentration mg/mL | Total (mg) | Activity
(m.Unit) |
Specific activity (Unit/g) | Total activity (m.Unit) | Purification (Fold) | yield | ||
| Crude Extract | 1500 | 3.41 | 5115 | 75.46 | 22.07 | 113190 | 1X | 100 |
| Ammonium Precipitation | 20 | 3.74 | 63.58 | 516.47 | 138.09 | 10329.4 | 6.51 X | 9.12 |
| DEAE Fast Flow | 62 | 0.164 | 10.153 | 122.257 | 746.589 | 7579.969 | 33.828 X | 6.69 |
| Sephacryl S- 200 | 60 | 0.077 | 4.62 | 92.43 | 1200.48 | 5546.25 | 54.368 X | 4.89 |
SDS-PAGE of crude and purified enzyme (Figure 2) showed that the purified enzyme still exhibited few band of proteins, this is suggested that either the enzyme is poorly purified or there are more than one protein showing lipase activity. Following separation by native gel and zymogram analysis (Figure 3 A,B) showed that there were four bands exhibiting lipase activity. This is suggested that there is more than one type of lipase produced by the isolate. There were few reports showing that lipase or esterase from microorganism could form as single or multiple protein22-23. In addition, some there are reports showed that single microorganisms expressed more than one type of lipases 24-25.
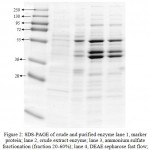 |
Figure 2: SDS-PAGE of crude and purified enzyme lane 1, marker protein; lane 2, crude extract enzyme; lane 3, ammonium sulfate fractionation (fraction 20-60%); lane 4, DEAE sepharose fast flow; lane 4, Sephacryl S-200 results.
|
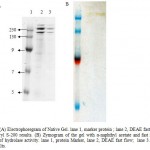 |
Figure 3 a: Electrophoregram of Native Gel. lane 1, marker protein ; lane 2, DEAE fast flow; lane 3, Sephacryl S-200 results. (B) Zymogram of the gel with α-naphthyl acetate and fast red TR for detection of hydrolase activity. lane 1, protein Marker, lane 2, DEAE fast flow; lane 3. Sephacryl S-200 results.
|
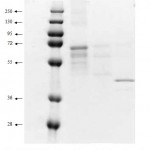 |
Figure 4: SDS-PAGE from electro elution result. lane 1, marker protein; lane 2, first band, lane 3, second and third bands, lane 4, the fourth band from native gel respectively.
|
Further analysis on the bands which have lipase activity were carried out by eluting each band from the native gel, the bands were then exposed to SDS-PAGE. The result showed that the fourth band exhibited single protein on the SDS-PAGE. Meanwhile, the other bands showed few protein bands but in similar pattern. This is suggested that the isolate expressed more than one type of lipases.
Acknowledgements
This research was partial supported by Graduate Research Grant to Fida Madayanti, and BPPS Scholarship and sandwich program to Febriani, Directorat General for Higher Education, Ministry of National Education Republic of Indonesia, and ITB Excelent Research Grant to Akhmaloka.
References
- Jaeger K. E. and Reetz M. T. Microbial Lipases Form Versatile Tools for Biotechnology, Trends Biotechnol, 16: 396-403 (1998).
- Yang J. and Yan J.S.Y. lip2, a novel lipase gene cloned from Aspergillus niger exhibits enzymatic characteristics distinct from its previously identified family member, Biotechnol Lett, 32: 951–956 (2010).
- Karadzie , Masui A., Zivkovic, L., and Fujiwara, N. Purification and Characterization of an Alkaline Lipase from Pseudomonas aeruginosa Isolate from Putrid Mineral Cutting oil as Component of metal working fluid, Journal of Bioscience and Bioengineering, 102: 82-89 (2006).
- Gupta R., Gupta N., and Rathi P. Bacterial Lipases: an Overview of Production, Purification and Biochemical Properties Appl Microbiol Biotechnol, 64: 763-781 (2004).
- Park D.S., Oh H.W., Heo S.Y., Jeong W.J., Shin D.H., and Bae, K.S. Characterization of an Extracellular Lipase in Burkholderia sp, The Journal of Microbiology, 45: 409-417 (2007).
- Kanwar S.S., Ghazi I.A., Chimni S.S., Joshi G.K., Rao G.K., Kaushal R.K., Gupta R., and Punj V. Purification and properties of a novel extra-cellular thermotolerant metallolipase of Bacillus coagulans MTCC-6375 isolate, Protein Expression and Purification 46: 421-428 (2006).
- Sharma R., Chisti Y., and Banerjee C. Production, Purification, Characterization, and Application of Lipase, Biotechnol. Adv., 19: 627 – 662 (2001).
- Kumar S., Kikon K., Upadhyay A., Kanwar, S.S., and Gupta. Production, Purification, and Characterization of Lipase from Thermophilic and Alkaliphilic Bacillus coagulans BTS-3 Protein Expression and Purification, 41: 38-44 (2005).
- Saxena R.K., Ghosh P.K., Gupta R., Sheda Dvidson W., Bradoo S., and Gulati R. Microbial Lipases, Potential Biocatalysts for the future Industry, Current Science, 77:101-115 (1999).
- Prazeres J.N.D., Cruz J.A.B., and Glaucia M. P. Characterization of Alkaline Lipase From Fusarium Oxysporum and The effect of Different Surfactants and Detergents on the Enzyme Activity, Brazilian Journal of Microbiology 37: 505-509 (2006).
- Salameh, M.A., and Wiegel.J. Purification and Characterization of Two Highly Thermophilic Alkaline Lipases from Thermosyntropha lipolytica, Applied and Environmental Microbiology 73: 7725-7731 (2007).
- Yohandini H., Madayanti, F., Aditiawati, P., and Akhmaloka. Diversity of Microbiol Thermophiles in A Neutral Hot Spring (Kawah Hujan A) of Kamojang Geothermal Field, Indonesia, Journal of Pure and Applied Microbiology, 2(2): 283-294 (2008).
- Baker G.C., Gaffar S.,Cowan D.A., and Suharto A.R. Bacterial community analysis of Indonesian hot springs, FEMS Microbiol. Lett., 200: 103-109 (2001).
- , Suharto A., Nurbaiti S, Tika I.N., and Warganegara F.M. Ribotyping Identification of Thermophilic Bacterium from Papandayan Crater, Proc. ITB Eng. Science, 38B: 1-10 (2006).
- Aditiawati P, Yohandini H, Madayanti F., and Akhmaloka. Microbial diversity of acidic hot spring (Kawah Hujan B) in geothermal field of Kamojang area, West Java, Indonesia The Open Microbiology J., 3: 58-66 (2009).
- Madayanti F., El Viera B.V., Widhiastuty M.P., and Akhmaloka. Characterization and Identification of Thermophilic Lipase Producing bacteria from Thermogenic Compost, Journal of Pure and Applied Microbiology, 2(2): 325-332 (2008).
- Widhiastuty M.P., Febriani, Yohandini H., Moeis M.R.,, Madayanti F., and Akhmaloka. Characterization and Identification of Thermostable Alkaline Lipase Producing Bacteria From Hot Spring around West Java, Journal of Pure and Applied Microbiology, 3(1), 27-40 (2009).
- Dussault,H.P. An Improved Technique for Staining Red-Halophilic Bacteria, Journal Bacteriol, 70: 484-485 (1955).
- Sambrook,J., and Fritsch E.F. Molecular Cloning A Laboratory Manual, 2nd Ed.Cold Spring Harbour Laboratory Press, New York, USA (1989).
- Lee, D.W., Koh, Y.S., Kim K.J., Kim B.C., Choi H.J., Kim., D.S., Suhartono M.T., and Pyun, Y.R. Isolation and Characterization of a Thermophilic Lipase from Bacillus thermoleovorans ID-1, FEMS Microbiology Letters, 179: 393-400 (1999).
- Bradford M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding, Biochem., 72: 248-254 (1976) .
- Kambourova, M, N., Kirilova, R., Mandeva A., Derekova. Purification and Properties of Thermostable Lipase from a Thermophilic Bacillus stearothermophilus MC7, Journal of Molecular Catalysis B, 22: 307–313 (2003).
- Kademi, N., Ait – Aldelkader, L. Fakhreddine J., and Baratti. Purification and Characterization of a thermostable esterase from the moderate thermophile Bacillus circulan, Appl Microbiol Biotechnol 54: 173-179 (2000).
- Salameh M.A and Wiegel.J. Purification and Characterization of Two Highly Thermophilic Alkaline Lipases from Thermosyntropha lipolytica, Applied and Environmental Microbiology, 73: 7727-7731 (2007).
- Fucin os, P., Domınguez, A., Sanroma, M.A., Longo,M..A., Ru a, M.L., and Pastrana, L. Production of Thermostable Lipolytic Activity, Prog, 21: 1198 -1205 (2005).

This work is licensed under a Creative Commons Attribution 4.0 International License.





