Manuscript accepted on : February 16, 2011
Published online on: 28-06-2011
Kapil D. Kamble
P.G. Department of Microbiology Sant Gadge Baba Amravati University, Amravati India.
ABSTRACT: Deoxyribonucleases (DNases) are the enzymes which break down the phosphodiester linkages of deoxyribonucleic acid (DNA) by addition of water and are categorized under hydrolases. They have large applications in almost all fields including therapeutic, industrial and genetic engineering fields. In present study Serratia marcescens strains were screened through a large number of soil samples. These strains were then studied for DNase production on DNase test agar and on the basis of zone of DNA hydrolysis around the colony; efficient two strains, a pigmented and non-pigmented strains were chosen further for comparative study. Attempt was to introduce a better DNase producer of the two strains to avail them for various applications. The morphological and biochemical properties were carried out and the phylogeny of this bacterium by 16S rRNA sequencing was studied in detail. The phylogenetic study and biochemical study confirmed that the strains belong to the genus Serratia marcescens. A quantitative estimation of the DNase was carried out which reveals that the non-pigmented strain is better DNase producing bacterium than the pigmented strain. The optimum conditions for both the strains were studied. Both strains produce the DNase optimally at 20°C and at pH 8. As both strains are isolated from soil; are non-pathogenic and can be handled with less care during the production. Further the pigmented Serratia marcescens produces prodigiosin a red colored pigment which may be responsible for lesser DNase productivity.
KEYWORDS:
DNase production; Phylogenetic study; pigmented and non-pigmented Serratia marcescens
Download this article as:| Copy the following to cite this article: Kamble K. D. Prediction of Better Deoxyribonucleic Acid Hydrolase Producing Bacterium among Pigmented and Non-pigmented Serratia marcescens Strains from Soil.. Biosci Biotech Res Asia 2011;8(1). |
| Copy the following to cite this URL: Kamble K. D. Prediction of Better Deoxyribonucleic Acid Hydrolase Producing Bacterium among Pigmented and Non-pigmented Serratia marcescens Strains from Soil.. Biosci Biotech Res Asia 2011;8(1).Available from: https://www.biotech-asia.org/?p=9360 |
Introduction
Nucleases are involved in replication, repair, restriction and recombination. These are also involved in transposition, transcription and topoisomerization, as well as RNA processing, RNA splicing, editing and interference (Mishra, 2002). The accumulation of 8-OHGua induces G:C to T:A transversion during DNA replication that is related to mutagenic events. The 8-OHGua is formed by Reactive Oxygen Species which is removed from DNA by endonucleases in DNA salvage pathways (Hui et al. 2003). Single strand specific endonuclease has been exploited in mutation studies (Tierney et al. 2005). In DNA foot printing also DNases are used which is a method of investigating the sequence specificity of DNA-binding proteins. This technique is used to study protein-DNA interactions both outside and within cells (www.wikipedia.org). DNases cleaves chromosomal DNA into nucleosomal fragments in 180-200 bp in the process of apoptosis and later on these fragments are broken into 20-30 nucleotides and then into single nucleotides (Sluyser Mels, 2005). Qin et al. (2005) has explored the feasibility of using capsid-targeted viral inactivation (CTVI) as an antiviral strategy against dengue infection and are also found useful against moloney murine leukemia virus. Derivatives of nucleotide products are prepared which mimics the viral activities (Howell et. al., 1988). Nucleases are also candidates for the enzymes producing a tasty substance, 5′-GMP. (Kobayashi et al. 2000). Nucleases are studied from plants, insects and fungi.
In present study Serratia marcescens strains were screened through a large number of soil samples. These strains were then studied for DNase production on DNase test agar and on the basis of zone of DNA hydrolysis around the colony; efficient two strains, a pigmented and non-pigmented strains were chosen further for comparative study.
Materials and Methods
Soil Sample Collection (Gary M. Banowetz et al., 2006)
Soil samples were collected using a 2.54-cm-diameter soil corer. The corer was cleaned after each sampling with water followed by methanol and samples were placed in zip-lock plastic bags and stored for no longer than 7 days at 6°C. (Gary M. Banowetz et al., 2006).
Screening of DNase Producing Bacteria (Schreier, J. B.1969)
Screening was done on DNase test agar with toludine blue. Toludine blue on reaction with DNA gives sky blue color. When DNA is hydrolyzed by bacterial action; toludine becomes free and being in a free form a faint pink or colorless halos is observed around the colony.
Morphological and biochemical characterization of efficient DNase producers.
Routine microbiological methods were adopted for this purpose.
Phylogenetic study of some efficient cultures (Saitou, N. and Nei, M. (1987), Felsenstein, J., (1985), Kimura, M. (1980), Tamura, et al., (2007).
Identification by 16S rRNA sequencing is the most preferred these days. The service was provided by Chromous Biotech Pvt. Ltd. Bangalore.
Estimation of DNase (Lachica R.V.F. et.al., 1972)
The characterized Serratia marcescens strains were grown in nutrient broth containing 0.2% DNA. The broth was centrifuged at 13,500 g to separate out the cells, so that supernatant is used as source of DNases as DNases are extracellular enzymes. Wells were then cut on methyl green DNA agar by cork borer of 2 mm diameter. A measured quantity (3ml) of DNase was added by micropipette aseptically. These were then added in the wells prepared. Smaller DNase conc. formed smaller zones and higher DNase conc. formed larger zones of DNA hydrolysis as seen in photo plate NO. 3 and 4. A standard graph already prepared was referred for quantitative estimation of DNase in the broth.
Result and Discussion
Screening of DNase producing bacteria
 |
Plate:1 Serratia marcescens on DNase testDNase Test agar with toludiene blue.
|
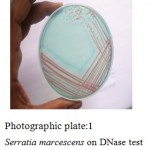 |
Plate:2 Serratia marcescens on DNase Test agar with toludiene blue.
|
Table 1: Morphological & Biochemical Characteristics of isolates from East region.
| Biochemical properties | Serratia marcescens | Serratia marcescens |
| (Pigmented) | (Non-pigmented) | |
| Gram staining | Gram – | Gram- |
| Motility | + | + |
| Indole | – | – |
| MR test | + | + |
| Voges-Proskauer | – | – |
| Citrate | + | + |
| Hydrogen sulfide | – | – |
| Motility | + | + |
| Lysine decarboxylase | + | + |
| Ornithine decarboxylase | + | + |
| Glucose: acid | + | + |
| Glucose: gasa | + | + |
| Lactose | – | – |
| Mannitol | + | + |
| Inositol | + | + |
| Arabinose | – | – |
| Raffinose | – | – |
| Rhamnose | – | – |
| Lipase (corn oil) | + | + |
| Cellobiose | + | + |
| Glycerol acid | + | + |
| Maltose | + | + |
| Glycerol gas | – | – |
| Gelatin | + | + |
| Pigment | + | – |
 |
Figure 1: The pigmented strain in above phylogenetic tree is 165 D.
|
 |
Figure 2: The non-pigmented strain in above phylogenetic tree is 165 E.
|
Phylogenetic study of the selected cultures
The pigmented strain in above phylogenetic tree is 165 D
The non-pigmented strain in above phylogenetic tree is 165 E
Estimation of DNase
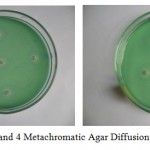 |
Plate 3: and 4 Metachromatic Agar Diffusion Assay of DNase.
|
Serratia marcescens a pigmented strain produced maximum DNase i.e. 6.1mg/ml at pH 9 and 6.25 mg/ml at temperature of 20°C. Other non-pigmented strain produced maximum DNase at pH 9 i.e. 6.3mg/ml and at temperature 30°C maximum DNase i.e. 6.35mg/ml was produced. This has shown that a non-pigmented strain of Serratia marcescens produces more DNase than pigmented strain.
References
- Desai, N. A. and Vepatu S., J. Biochem., 267: 5123-5135 (2000).
- Deshmukh, S. S. and Vepatu S., Federation of European Microbial Society, 237(2): 273-278 (2004).
- Felsenstein, J., Evolution 39: 783-791(1985).
- Gary, M. B. et al., Environ Qual 35 : 133-140 (2006).
- Howell, H. G. et al., Org. Chem. 53: 85-88 (1988).
- Hui, H. Z., Jeong Y.C. and Myung H.C., Chinese Chemical Letters 14(5): 529-532, (2003).
- Jeffries, C. D., Hoffman, D. F. and Guse, D. G., J. Bacteriol. 73, 590 – 591(1957).
- Kimura, M., Journal of Molecular Evolution 16, 111-120, (1980).
- Kobayashi, H. et.al., Biosci. biotechnol. biochem. 64(5), 948-957 (2000).
- Lachica, R. V. F., Hoeprich, P. D. and Franti, C. E., Environ. Microbiol. 24(6), 920– 923 (1972)
- Mishra, N.C. Hoboken, N.J., USA John Wiley and Sons.189-190, (2002).
- Qin, C.F., Qin E., Yu, M., Chen, S.P., Jiang, T., Deng, Y, Q., Duan, H.Y. and Zhao, H., Archives of Virol. 150 (4), (2005).
- Rangrajan, S., Shankar, V., Biochim Biophys Acta. 1473 (2-3), 293 – 304, (1999).
- Saitou, N. and Nei, M., The neighbor-joining method . Molecular biology and Evolution 4406-425, (1987).
- Schreier, J. B., Am. J. Clin. Pathol. 51711-716, (1969).
- Smith, P. B., Hancock, G. A. and Rhoden, D. L., microbiol. 18 (6), 991-993, (1977).
- Tamura, K., Dudley, J., Nei, M. and Kumar, S., Molecular Biology and Evolution. 24, 1596-1599 (2007).
- Tierney, M.B. and Lamour, K.H., The Plant Health Instructor. 1025-01, (2005).
- Verma, K. P., Saini, A., Chopra A. K., Chakraborti P. K., Singh Y. and Choudhary S., Nucleic acid Res. 33 (8), 2707 (2005).
- Ying, Guo-Qing, Lu-E shi, Yi Yu, tang Zehn-Xing, Chen Jian Shu., Process Biochemistry, 41, 1276-1281, (2006).

This work is licensed under a Creative Commons Attribution 4.0 International License.





