Manuscript accepted on : 19 February 2012
Published online on: --
Molecular Screening of Virulence Genes from Salmonella enterica Isolated from Commercial Food Stuffs
Arunava Das1*, S. Sree Hari1, U. Shalini2, A. Ganeshkumar3 and M. Karthikeyan4
1Department of Biotechnology, Bannari Amman Institute of Technology, Sathyamangalam - 638 401, India.
2Centre for Biotechnology, A. C. Tech Campus, Anna University, Chennai - 600 025, India.
3Department of Biological Sciences, BITS, Pilani - K. K. Birla Goa Campus, Zuarinagar - 403 726, India.
4Biocon Limited, Electronic City, Bangalore - 560 100, India.
Corresponding Author E-mail: drarunavadas@rediffmail.com
ABSTRACT: The present research work was carried out for the screening of virulence genes associated with the Salmonella enterica isolated from commercial food stuffs by polymerase chain reaction (PCR). A total of 134 samples of commercial food stuffs constituting of raw meats of poultry, pork, beef, raw eggs, dairy and bakery products were purchased from the departmental stores, supermarkets and local butcher shops of Salem, Erode and Coimbatore districts of Tamil Nadu, India. Samples were aseptically processed for the isolation of S. enterica through broth enrichment methods. PCR was performed with various virulence genes specific primers of S. enterica. Microbiological investigations resulted in Salmonella isolates in 35 (26.11%) samples. In PCR, invasive gene (invA; 244bp), Salmonella enterotoxin gene (stn; 617bp), plasmid encoded fimbriae (pefA; 700bp), Salmonella Enteritidis fimbriae (sefC; 1103bp) and Salmonella plasmid virulence C gene (spvC; 571bp) were detected in 100%, 100%, 51.42%, 25.71% and 42.85% isolates respectively. Present study suggested that invA and stn virulence genes are much conserved in S. enterica isolated from commercial food stuffs and could be used independently as a gene marker for the rapid detection of the virulent strains of S. enterica. The prevalence of spvC gene is restricted into the isolates of a few definite sources. The result emphasized the risk of transferring these zoonotic organisms to human via food chain is impending danger for the mankind.
KEYWORDS: Salmonella enterica; commercial food stuffs; PCR; virulence
Download this article as:| Copy the following to cite this article: Das A, Hari S. S, Shalini U, Ganeshkumar A, Karthikeyan M. Molecular Screening of Virulence Genes from Salmonella enterica Isolated from Commercial Food Stuffs. Biosci Biotech Res Asia 2012;9(1) |
| Copy the following to cite this URL: Das A, Hari S. S, Shalini U, Ganeshkumar A, Karthikeyan M. Molecular Screening of Virulence Genes from Salmonella enterica Isolated from Commercial Food Stuffs. Biosci Biotech Res Asia 2012;9(1). Available from: https://www.biotech-asia.org/?p=9703 |
Introduction
The genus Salmonella is an enteric, gram negative, rod shaped bacteria grouped in the family of Enterobacteriaceae and one of the most important food-borne pathogens associated with a hyperendemic diarrhoeal disease called Salmonellosis around the world affecting both man and animal alike (Prakash et al., 2005). Salmonellosis is a fatal disease and is spread to human being from eating (orally) of improperly cooked foods such as meat, eggs, unpasteurized milk, dairy products, bakery products and also by direct contact with faeces/diarrhoea from infected animals (Holt et al., 1994).
Salmonella have several sub species among them Salmonella enterica subspecies enterica (subspecies 1) is responsible for 99.5% of food borne illness in humans and animals (Pignato et al., 1998). S. enterica includes several virulence genes which encode products that assist the organisms to express its virulence in the host. Among the virulence genes, inv, sef and pef are considered for adhesion and invasion of the pathogen in the host system and spv gene for systemic disease state in the host cells. While stn virulence gene codes for enterotoxin production sop and pip genes are associated actual expressions of host pathogenic processes (Murugkar et al., 2003). Hence, accurate and systematic method must be adopted for the screening of virulence genes from S. enterica isolates originated from the infected samples.
Commercial food products mainly constituted by poultry and poultry based products have been implicated as a major reservoir of Salmonella infections in humans which are predominantly considered under public health status and economic value (Salehi et al., 2005). Therefore, the prevention of Salmonella infection via commercial food products should be given primary importance by the food processing industries (Mahé et al., 2008). Present study was undertaken to isolate S. enterica from the commercial food stuffs and to screen its virulence genes present within the isolates by PCR for epidemiological study.
Materials and Methods
Sample details
A total of 134 commercial food stuffs samples including raw meats of poultry meat (32), pork meat (18), beef meat (18), raw eggs (25), dairy products (26), and bakery products (15) were collected from randomly selected departmental stores, supermarkets and local butcher shops in Salem, Erode and Coimbatore districts, Tamil Nadu, India. The food stuffs were purchased in regular consumer bags and immediately transferred to the laboratory for microbial investigation.
Isolation of Salmonella enterica serovars
Food samples (1g) were placed in 10 ml of buffered peptone water (Hi-Media, Mumbai) as pre-enrichment media, and incubated at 370C for 18 h. The incubated broth samples were enriched by transferring 0.1 ml of the broth into 1 ml tetrathionate (TT) broth (Hi-Media, Mumbai), incubated at 370C for 24 hr. The cultured broths were streaked onto xylose lysine deoxycholate agar (Hi-Media, Mumbai) and brilliant green agar (Hi-Media, Mumbai), incubated at 370C for 24 hour. Suspected colonies of Salmonella were purified and identified by motility test, Gram and flagellar staining, indole test, methyl red test, Voges-Proskauer test, citrate test (growth on Simmon’s citrate agar), urease test, gelatin hydrolysis, H2S production, acid and gas production test from glucose, mannitol, maltose, sorbitol, adonitol, sucrose, salicin, and lactose.
Screening of virulence genes by duplex and single step PCR
Suspected bacteria were screened for the detection of virulence encoding genes like the invasion gene (invA), Salmonella plasmid virulence gene (spvC), plasmid encoded fimbriae (pefA), Salmonella Enteritidis fimbriae (sefC) and Salmonella enterotoxin gene (stn) by PCR. In this study, two sets of duplex PCR and one set of single step PCR were performed. The first set of dPCR was used for the amplification of invA and spvC genes, while, second set of dPCR performed for the detection of pefA and sefC genes. In single step PCR, amplification was carried out for the stn gene. The specific forward and reverse primer pairs (Table 1) were commercially synthesized (Eurofins Genomics India Pvt. Ltd., Bangalore). S. enterica serovar Typhimurium (MTCC 98) and Aeromonas hydrophila (MTCC 646) strains were used as positive and negative controls respectively.
Preparation of template DNA
Bacterial colonies were freshly grown in nutrient agar plates, suspended in 150μl of sterile distilled water in a micro centrifuge tube, gently vortexed and boiled for 10 min in a water bath. Micro tubes were then centrifuged at 10000 rpm for 5min at 40C. Top supernatant were carefully collected and used as a source of template DNA.
PCR conditions
In every case, the PCR amplifications was carried out in 25μl reaction volume in Thermal Cycler (Eppendorf, Germany) containing 12.5μl of 2 × PCR master mix (Promega, USA) containing 4mM magnesium chloride, 0.4mM of deoxynucleotide triphosphates (dNTPs), 0.5U of Taq DNA polymerase, 150mM tris-hydrochloric acid, pH 8.5 (Promega, USA), 2.5μl of template DNA and a specific concentration of each forward and reverse primer pairs (Table 1).
In each set, PCR had an initial denaturation for 4 min at 940C and a final extension for 7 min at 720C. The amplification cycles for the first set of dPCR undergone denaturation at 940C for 30 sec, annealing at 560C for 30 sec and extension at 720C for 2 min respectively. The second set of dPCR was run for the denaturation at 940C for 55 sec, annealing at 550C for 55 sec and extension at 720C for 55 sec respectively. The single step PCR was exclusively run for the denaturation at 940C for 60 sec, annealing at 590C for 60 sec and extension at 720C for 60 sec respectively.
Agarose gel electrophoresis
The PCR amplicons (5μl) were electrophoresed in 1.2 % agarose gel in TAE (Tris-acetate-EDTA, pH 8) buffer, stained with ethidium bromide (0.4 µg / ml) and observed under Gel Documentation system (Universal Hood, BIORAD, Italy).
Results and Discussion
In the present study, prevalence of Salmonella enterica was made possible by broth enrichment method of isolation from various sources and geographical locations. A total of 35 (26.11%) S. enterica were isolated from 134 commercial food samples (Table 2). Among these 35 isolates, 12 (37.5%), 6 (33.33%), 5 (27.77%), 6 (24.00%), 5 (19.23%) and 1 (6.25%) were isolated from poultry meat, beef meat, pork meat, raw eggs, dairy products and bakery products respectively. The isolated Salmonella bacteria were found to be motile, Gram negative, flagellated, indole negative, methyl red positive, Voges-Proskauer negative, citrate positive, urease negative, gelatin hydrolysis negative, positive for H2S production and acid and gas production from glucose, mannitol, maltose, and sorbitol. No acid production was observed from adonitol, sucrose, salicin, and lactose. Similar biochemical characteristics from S. enterica serovar Typhi isolated from humans were observed (Ganeshkumar et al., 2010). Microbiological investigation showed that the isolation frequencies of S. enterica were higher in meat samples than raw eggs, dairy and bakery products. This also implied that poultry, beef and pork meat, raw eggs and dairy products provide suitable environment for S. enterica to grow at an increasingly higher rate irrespective of its geographical locations. In other study, the prevalence of Salmonella in chickens and beef cattle was found to be 32.5% (39/120) and 4.6% (11/240) respectively and among the cattle isolates, the prevalence rate was found significantly higher in fasted cattle (7.46%), than in non-fasted cattle (0.94%) (Abouzeed et al., 2000). Wegener et al., (1997) also detected high frequencies of Salmonella isolates from poultry, poultry products, cattle and dairy products and suggested that these food products are the major sources of Salmonella infections in humans.
The virulence of Salmonella is linked to a number of virulence factors and in this study; few of them were screened by PCR. In the first set of dPCR which run for invA (244bp fragment) and spvC (571bp fragment) genes resulted positive amplifications in 35 (100%) and 15 (42.85%) isolates respectively (Table 2, Figure 1), similarly, the second set which run for pefA (700bp fragment) and sefC (1103bp fragment) genes showed positive amplifications in 18 (51.42%) and 9 (25.71%) isolates respectively (Table 2; Figure 2). The single step PCR which performed for stn (617bp fragment) gene resulted positive amplifications in all the 35 (100%) isolates (Table 2, Figure 3). The detection of invA, spvC, pefA, sefC and stn genes from the isolates of S. enterica originated from food, faecal or clinical samples by PCR were investigated by various researchers (Swamy et al., 1996 & Chiu and Ou, 1996 & Shome et al., 2006).
Table 1: Details of virulence gene specific primers used in PCR.
| Virulence genes | Oligonucleotides (5/ to 3/) | Concentration (µM) | Fragment size (bp) | Reference |
| invA | For:acagtgctcgttacgacctgaat Rev:agacgactggtactgatcgataat | 1 | 244 | Chiu and Ou, (1996 ) |
| spvC | For:actccttgcaccaaccaaatgcgga Rev:tgtcttcttgcatttcgccaccatca | 571 | ||
| pefA | For:tgtttccgggcttgtgct
Rev:cagggcatttgctgattcttcc |
0.5 | 700 | Murugkar et al., (2003) |
| sefC | For:gcgaaaaccaatgcgactgta
Rev:cccaccagaaacattcatccc |
1103 | ||
| stn | For:ttgtgtcgctatcactggcaacc Rev: attcgtaacccgctctcgtcc | 1 | 617 |
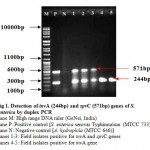 |
Figure 1: Detection of invA (244bp) and spvC (571bp) genes of S. enterica by duplex PCR.
|
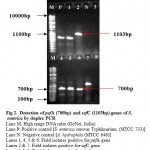 |
Figure 2: Detection of pefA (700bp) and sefC (1103bp) genes of S. enterica by duplex PCR.
|
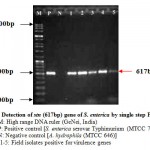 |
Figure 3: Detection of stn (617bp) gene of S. enterica by single step PCR
|
In the present study, both invA and spvC genes were positive for 15(42.85%) isolates, while only 20(57.14%) isolates were positive for invA gene alone. In a study conducted with 38 isolates of Salmonella serovars, all 38 were found positive for invA gene whereas, 16 isolates produced additional spvC genes (Chiu and Ou, 1996). Swamy et al., (1996) reported that out of 245 Salmonella isolates, invA genes were amplified by all and 37 (15.1%) isolates mostly originated from egg contents or from the egg production environment harbored spvC genes in addition to invA genes. Montenegro et al., (1991) found that frequencies of detection of spvC gene vary from 48 to 87% within the S. enterica isolated from faecal, food or environmental samples. The detection frequency of spvC gene in this study was found to be 42.85% from the isolates originated from only meat and raw egg environments. This suggested that the prevalence of Salmonella virulence plasmids is restricted within the isolates of some defined origin of sources.
In this study, none of the S. enterica isolates amplified pefA and sefC genes together in dPCR, all the pefA and sefC genes were independently detected. Like invA gene, stn gene also demonstrated 100% amplification among the isolates. The PCR result suggested that the distribution of pefC gene was more in meat samples compared to the dairy products and raw eggs, but, sefC gene was amplified higher in isolates originated from raw eggs. There was only one isolate originated from the bakery products and was found negative for pefA, sefC and spvC genes. In a comparable study for the distribution of stn, pef and sef genes among 95 Salmonella isolates originated from poultry, pig, cattle and humans samples, all 95 (100%) were found to harvour stn gene, whereas, 85 (89.47%) and 36 (37.89%) isolates were positive for pef and sef genes respectively (Murugkar et al., 2003). In another study carried out with 23 S. enterica isolates originated from faecal samples of pigs, and poultry, all were found positive for stn genes, whereas, 11 and 4 isolates were positive for pefA and sefC genes respectively (Shome et al., 2006).
The 100% detection rate of inv and stn genes by PCR suggested that these two genes are conserved among S. enterica isolates (De Oliveira et al., 2003 & Murugkar et al., 2003). The virulence genes screening by PCR also suggested that both inv and stn genes are predominant virulence genes necessary for the serovars of S. enterica to express virulence in the host. The PCR assays employing with either invA or stn virulence genes appeared to be rapid, sensitive, and specific means to identify S. enterica isolates from commercial food stuffs.
Although, commercial food samples in this study were collected randomly from Coimbatore, Erode and Salem Districts of Tamil Nadu, the number of Salmonella isolated from each geographical location varied. From Table 2, it is very clear that the isolation rate of Salmonella were much higher in the samples obtained from Coimbatore 16 (45.71%), than Erode 13 (37.14%) and Salem 6 (17.14%). This study gives a preliminary idea about the distribution of S. enterica isolates in the three neighbouring districts located in the Southwest of the Chennai (state capital), Tamil Nadu. The persistence of Salmonella in the commercial food environment was an important characteristic in its prevalence. The PCR detection of invA, spvC, pefA, sefC and stn genes among the isolates of S. enterica also signified the higher risk of food-borne infections caused by these zoonotic bacteria into humans. Therefore, aseptic storage practice and proper sterilization of food products needs to be improved in the departmental stores, supermarkets and even in local butcher shops in Tamil Nadu India.
Table 2: Isolation and virulence gene detection of S. enterica isolates.
| Source of sample collection | No of samples collected from various geographical locations | No of Salmonella isolated from various geographical locations | No of Salmonella isolates / no of sample tested (%) | No of isolates positive for virulence genes by PCR | ||||
| invA | spvC | pefA | sefC | stn | ||||
| Poultry meat | 12b
10a 10c |
5b
2a 5c |
12/32 (37.5) | 12 | 6 | 8 | 1 | 12 |
| Pork meat | 7a
6b 5c |
1a
2b 2c |
5/18 (27.77) | 5 | 1 | 3 | 1 | 5 |
| Beef meat | 8c
6b 4a |
2c
3b 1a |
6/18 (33.33) | 6 | 2 | 4 | 1 | 6 |
| Raw eggs | 10c
7a 8b |
3c
1a 2b |
6/25 (24.00) | 6 | 6 | 1 | 5 | 6 |
| Dairy products | 10c
9b 7a |
3c
1b 1a |
5/26 (19.23) | 5 | 0 | 2 | 1 | 5 |
| Bakery products | 6c
6b 3a |
1c
0b 0a |
1/16 (6.25) | 1 | 0 | 0 | 0 | 1 |
| Total (%) | 134 [38(28.35)a, 47(35.07)b, 49(36.56)c] | 35 [6(17.14)a, 13(37.14)b, 16(45.71)c] | 35/134 (26.11) | 35 (100) | 15 (42.85) | 18 (51.42) | 9 (25.71) | 35 (100) |
a: Salem, b: Erode, c: Coimbatore
Conclusion
Present study suggested that PCR is a sensitive, reliable tool for the rapid detection of S. enterica serovars from the commercial food products. High prevalence of virulent strains of S. enterica from poultry, beef and pork meat products, raw eggs and dairy products clearly indicated that consumption of improperly cooked commercial food stuffs are unsafe and may result in serious food-borne illness in humans. Further, molecular analysis need to be carried out to achieve the accurate distribution of the virulence genes among the various serovars of S. enterica and their protein products in the molecular pathogenesis in humans as well as animals which will help us to develop preventive measures against the deadly pathogens.
Acknowledgements
All the authors gratefully acknowledge the management of Bannari Amman Institute of Technology, Sathyamangalam for providing wonderful ambience to carry out research work.
References
- Prakash, B., Krishnappa, G., Muniyappa, L. and Santhosh, K.B. Epidemiological characterization of avian Salmonella enterica serovar infections in India. J. Poult. Sc., 2005; 4: 388-395.
- Holt, P.S., Buhr, R.J., Cunningham, D.L. and Porter, R.E. Effect of two different molting procedures on a Salmonella enteritidis Poult. Sc., 1994; 73: 1267-1275.
- Pignato, S., Giammanco, G., Santangelo, C. and Giammanco, G. Endemic presence of Salmonella bongori 48:Z35: Causing enteritis in children in Sicily. Microbiol., 1998; 149: 429-431.
- Murugkar, H.V., Rahman, H. and Dutta, P.K. Distribution of virulence genes in Salmonella serovars isolated from man and animals. Indian J. Med. Res., 2003; 117: 66-70.
- Salehi, Z., Mahzounieh, M. and Saeedzadeh, A. Detection of invA gene in isolated Salmonella from broilers by PCR method. J. Poult. Sc., 2005; 4: 557-559.
- Mahé, A., Bougeard, S., Huneau-Salaün, A., Le Bouquin, S., Petetin, I., Rouxel, S., Lalande, F., Beloeil, P.A. and Rose, N. Bayesian estimation of flock-level sensitivity of detection of Salmonella, Enteritidis and Typhimurium according to the sampling procedure in French laying-hen houses. Prev. Vet. Med., 2008; 84: 11-26.
- Ganeshkumar, A., Shalini, U. Vaishnavi, K., Remya I., Lakshmanaswamy, A., Vasanthi, N.S. and Das A. Rapid detection of Salmonella enterica serovar Typhi from humans. Pure Appl. Microbiol., 2010; 4: 837-841.
- Abouzeed, Y.M., Hariharan, H., Poppe, C. and Kibenge, F.S. Characterization of Salmonella isolates from beef cattle, broiler chickens and human sources on Prince Edward Island. Immunol. Microbiol. Infect. Dis., 2000; 23: 253-266.
- Wegener, H.C., Hald, T., Wong, L.F., Madsen, M., Korsgaard, H., Bager, F., Gerner-Smidt, P. and Molbak, K. Salmonella Control Programs in Denmark. Infect. Dis., 2003; 9: 774-780.
- Swamy, S.C., Barnhart, H., Lee, M.D. and Dreesen, D.W. Virulence determinants invA and spvC in Salmonellae isolated from poultry products, wastewater and human sources. Environ. Microbiol., 1996; 62: 3768-3771.
- Chiu, C.H. and Ou, J.T. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth–multiplex PCR combination assay. Clin. Microbiol., 1996, 34: 2619-2622.
- Shome, B.R., Rahman, H., Shome, R., Murugkar, H.V., Mazumder, Y. Das, A., Kumar, A. and Bujarbaruah, K.M. Detection of virulent genes and genotyping of Salmonella enterica isolated from livestock and poultry. Indian Vet. J., 2006; 83: 934-938.
- Montenegro, M.A., Morelli, G. and Helmuth, R. 1991. Heteroduplex analysis of Salmonella virulence plasmids and their prevalence in isolates of defined sources. Pathogen. 11: 391–397.
- De Oliveira, S.D., Rodenbusch, C.R., Michael, G.B., Cardoso, M.I.R., Canal, C.W. and Brandelli, A. Detection of virulence genes in Salmonella enteritidis isolated from different sources. Brazilian J. Microbiol.2003; 34: 123-124.

This work is licensed under a Creative Commons Attribution 4.0 International License.





