How to Cite | Publication History | PlumX Article Matrix
Thirumala Mothe 1* , Patnam Umashankar 2
, Patnam Umashankar 2 and Vishnuvardhan Reddy Sultanpuram3
and Vishnuvardhan Reddy Sultanpuram3
1Department of Applied Biosciences, Microbial Ecology Laboratory, UCS&I, Mahatma Gandhi University, Anneparthy,Yellareddygudem (PO), Nalgonda-508254, Telangana, India.
2Bioanalytical Department, Axis clinicals, Hyderabad.
3Microbztech Lab Pvt. Limited, Plot No. 102, Phase-I, Sri Sai Dwarakapuri Colony, Cherlapally, Nalgonda-508001, Telangana, India.
Corresponding Author E-mail: thirumala_21@yahoo.com
DOI : http://dx.doi.org/10.13005/bbra/2900
ABSTRACT: Consuming of water contaminated with high fluoride concentration for a very long time causes health problems such as, dental and skeletal fluorosis. Hence, defluoridation of water is essentially required before consumption, when water is contaminated with high fluoride concentration. In this present study, research was focussed on to isolate bacteria which are showing fluoride resistantance from samples of ground water from high fluoride affected regions of Nalgonda. After analysis of 10 samples from different areas of Nalgonda district, two samples of ground water from Narketpally and Nampally showed the high fluoride concentrations of 9.18 ppm and 7.55 ppm respectively. Hence, in the present study, Narketpally ground water sample with highest ppm was considered to isolate fluoride resistant bacteria. A total of eight fluoride resistant organisms were purified from this sample with varying fluoride resistance on Luria Bertani agar with varying fluoride concentraion from 25mg/L to 600mg/L at pH 7. Among the eight strains isolated, three strains MB1, F and G were showing high fluoride resistance (up to 500mg/L), which were further explored for their role in bioremediation of fluoride. In batch mode study, MB1 strain showed high fluoride degradation of 68%, whereas, F and G strains showed 57% and 44%fluoride removal, respectively, when fluoride concentration was present at 20 mgL-1 at 30 °C temperature and pH 7, with dextrose (10 g) utilised as source of carbon per 100 mL media after incubation of 8 days. Results indicate that, MB1 possibly a potential fluoride resistant bacterium with high fluoride bioremediation capacity.
KEYWORDS: Bioremediation of Fluoride; Fluoride Resistant Bacteria; Nalgonda District
Download this article as:| Copy the following to cite this article: Mothe T, Umashankar P, Sultanpuram V. R. Isolation of Fluoride Resistant Microorganisms from Fluoride Contaminated Ground Water Samples of Nalgonda district and their role in Bioremediation. Biosci Biotech Res Asia 2021;18(1). |
| Copy the following to cite this URL: Mothe T, Umashankar P, Sultanpuram V. R. Isolation of Fluoride Resistant Microorganisms from Fluoride Contaminated Ground Water Samples of Nalgonda district and their role in Bioremediation. Biosci Biotech Res Asia 2021;18(1). Available from: https://bit.ly/3dAoM74 |
Introduction
Fluorine is one amongst the copious elements on earth as well as, acts as a major environmental pollutant arising from usual as well as industrial sources (Whitford GM.,1983). The fluoride ion (F) concentration in the surface water is day by day increasing withquick industrialization and pollution with some insecticides (Aguirre-Sierra A et al., 2013). Ground water samples of greater than twenty countries including India have reported increased concentrations of Fluoride in them (Meenakshi and R.C.Maheshwari, 2006). TheWHO has recommended a maximum allowed concentration (MAC) of F–in source of drinking water is 1.5 mg/L, above which health problems are common. (WHO, 1984)
The fluoride ion acts as a protoplasmic toxin with which many biochemical reactions can get effected inside living cells, when present even at low concentrations (Erenet al., 2005). Fluoride stimulates oxidative damage by producing reactive oxygen species inside the cells which changes intracellular redox homeostasis (Podderet al., 2010 and Chattopadhyay et al., 2011) Fluoride binds with DNA resulting ininjury to DNA which possibly causes chemical carcinogenesis (Zhan et al., 2006; Podderet al., 2008;Choubisa, 2012).
One of the important concern with respect to environment nowadays is the escalating levelof fluoride in resources of groundwater (Susheela, 2013). These fluoride deposits are causing fluorosis in 17 states of India (UNICEF, 1999). Most of the researchers worked on fluoride concentration in ground water of Nalgonda district had reported high concentrations (Ayoob and Gupta, 2006; Ibrahim, 2011).
Increased levels of fluoride can affect the growth of microbes, their activity and organic matter degradation. (Zang et al., 2019) Hence, fluoride can be used as an additive in toothpastes to inhibit the formation of caries (Liao Y et al., 2017).
During evolution, microorganisms attained various abilities to resist certain concentrations of fluoride (Maltz and Emilson., 1982) as the result of genomic modifications (Liao Y et al., 2015; Mitsuhataet al., 2014).With the help of different mechanisms like metal sorption, oxidoreduction reactions and pumping out toxic elements present in them, microorganisms can overcome the unsuitable environments and use the various hazardous elements (Banerjee et al., 2016). To pump out fluoride ions some microorganisms consists of Fluoride antiporters with which they export these ions out of the cell and import protons, which causes the reduction in intracellular level of fluoride. Hence, mutations in genes related to fluoride antiporters decreased the resistance of S. mutans to fluoride(Liao Y et al., 2018).
Fluoride resistant microorganisms play a significant function in bioremediation of fluoride and convert it as less hazardous type (Chouhan et al., 2012) However, studies regarding the bioremediation of Fluoride are rare. In this paper,a study about fluoride-resistant bacterial strainsisolated from Fluoride contaminated ground water of Nalgonda district and analysis of their bioremediation capability was reported.
Materials and Methods
Sample Collection
Ground water samples were brought from water pumps and electric bore wells of fourteen different locations in Nalgonda district. For sample collection, bottles of 20mL capacity were washed rigorously with soap water and 8M HNO3 followed by distilled water. Before collecting samples from the bore wells or hand pumps,water was pumped out from them for about 5 to 10 min to get rid of standing water from the pipe lines of the same. Sample bottles were cleaned with the samples of ground water before final sample collection.
Fluoride estimation and pH analysis of water samples
Thesamples of ground water were collected from various places in Nalgonda disctrict were analysed for fluoride levelsusing a Fluriode ion selective electrode after calibration. In this test,1 mL of TISAB III (Total ionic strength adjustment buffer III solution)reagent was carefully mixed with 10 mL of ground water sample and measured using the fluoride ion selective electrode. The ground water samples which were brought from various sites in Nalgonda district were also tested with pH meter to know their pH profile.
Isolation and adjustment of bacterial isolates on media with Sodium Fluoride
All the glassware was rinsed thoroughly with 10 % HCL before use. High fluoride containing samples of ground water were used for serial dilution and then plated on to Luria–Bertani (LB) agar medium consisting of (g/L-1) Casein Enzyme Hydrolysate (Tryptone) (10), Yeast Extract (5),NaCl (5g),Agar (15). Plates were then incubated at pH 7.0±0.2 and temperature 30°C for 24 h. Bacterial isolates were arbitrarily choosen and purified. After purification of bacterial strains on LB agar, each and every strain was plated on to media with various sodium fluoride (NaF) amounts(25mg/L,50mg/L,100mg/L,200mg/L, 300mg/L, 400mg/L, 500mg/L and 600mg/L)in stepwise manner for their adaptation and resistance study.
Initially, inoculation of LB broth consisting of 25mg/L sodium fluoride (as in the plate medium)was done with Fluoride resistant micro organisms and incubated at 30° C, 120 rpm for 24 h. Sudden exposure of isolates to the high concentrations of fluoride may inhibit the growth of the same hence low concentrations of fluoride was initially selected to let the micro-organisms to get adapted to the fluoride containing media. The isolates were then exposed to a high fluoride concentration on Luria Bertani agar plates with 50 mg/L sodium fluoride concentration. After incubation, the same bacteria were then grown in LB broth consisting of 50 mg/L of sodium fluoride. The same approach was redone for the media with till 600 mg/L fluoride concentration. After three subcultures, Fluoride-resistant isolates were selected from the LB-NaF agar plates.
Characterization of selected bacterial isolates
Morphological, physiological and biochemical characteristics of the high fluoride resistant three isolates (MB1, F and G) were observed in this study. In morphological characteristics, colony morphology and gram’s test were performed.
Fluoride resistant organisms were tested for optimum pH and temperature for their optimum growth using LB broth with respective NaF concentration. Growth of the purified fluoride resistant organisms were checked in LB broth with respective NaF concentration at different temperatures i.e. 5°C to 45°C with every 5°C increment in an incubator for 48h. Growth ofthe purified fluoride resistant organisms was checked in LB broth consisting of respective NaF concentration at different pH, ranging from 2 to 12 in an incubator for 48hat 30 °C.
Biochemical tests performed on fluoride resistant organisms were to test their protease and amylase producing capabilities. To understand the capacity of protease enzymatic activities of the purified strains, they were first grown on skim milk agar (skim milk powder 28g/L; Yeast extract 2-5g/L; Dextrose 1g/L; Agar 25g/L; distilled water 1L) for 48 hours and observed for proteolysis activity. Fluoride resistant organisms with protease activity hydrolyzed casein and formed clear zone encircling the colonies.
To test the Amylase activity of the purified fluoride resistant isolates, first they were grown on starch agar media (peptone 5g /L; meat extract 3g /L;starch 2g/L; agar 25g/L; distilled water 1L) for 48h and plates were flooded with iodine solution (Iodine crystals 0.34g; KI 0.66g; distilled water 100mL). Amylase positive isolates formed a transparent halo area encircling the colony. The transparent halo area diameter is directly proportional to the starch hydrolyzing activity of the particular strain under study.
Determination of Bioremediation of fluoride by the selected three high fluoride resistant strains
To determine the fluoride removal activity of the three high fluoride resistant strains ( MB1, F and G), they were inoculated and incubated at temperature 30°C and pH 7 in 100 mL LB broth with 20 mg/L fluoride concentration (sodium fluoride (NaF)) in 250 ml conical flasks at 120 rpm with dextrose (10 g) as carbon source. Fluoride content was determined on every second day till incubation of 8 days. After which there was no considerable reduction in fluoride concentration, which must be due to bacterial cell count reduction. Maximum fluoride concentration observed in fluoride contaminated water was 19.2 mg/L, hence broth with 20 mg/L fluoride concentration was selected for the test. Sample (15ml) was taken out from the broth on every other day (i.e. days 0, 2, 4, 6, 8 and 10),during incubation of 10 days and centrifuged at 4500 rpm for 15 min, and then supernatant was tested for analyzing fluoride concentration with fluoride ion selective electrode(Mukherjee, S., et al., 2017).The above test was performed in triplicate.
Results and Discussion
Fluoride Estimation and pH analysis of water samples
Ten samples brought from different sites of Nalgonda district were analyzed for their fluoride concentration using fluriode ion selective electrode (Figure 1) and their pH (Table 1). Narketpally ground water sample was showing the highest Fluoride concentration i.e. 9.18 ppm, which was selected for isolation of Fluoride resistant bacteria.Whereas, Ramdas thanda (Nampally) ground water sample has less Fluoride concentration of 1.55 ppm.
 |
Figure 1: Thermo Scientific ORION STAR A214 ISE meter used for Fluoride estimation of ground water samples. |
Table 1: Fluoride concentration and pH values of the ten collected samples from ground water in Nalgonda district.
| Sample No | Location | Sample pH | Concentration of F– (ppm) |
| 1 | Narketpally | 8 | 9.18 |
| 2 | Chandur | 7 | 1.96 |
| 3 | Bangarigadda (Chandur) | 7 | 4.88 |
| 4 | Nermata (Chandur) | 8 | 5.61 |
| 5 | Ramdas thanda (Nampally) | 7 | 2.23 |
| 6 | Ramdas thanda (Nampally) | 8 | 1.55 |
| 7 | Peddapuram (Nampally) | 7 | 1.60 |
| 8 | Nerellapally (Nampally) | 7 | 7.55 |
| 9 | PadamatithallaGudem (Chandur) | 8 | 6.24 |
| 10 | MadaYadavally (Narketpally) | 7 | 6.55 |
Adjustment of bacterial isolates on media with Sodium Fluoride
In adaptation and resistance study, a total of eight fluoride resistant strains (MA, MB, MB1, MB1, D2, D1, E1, F and G) were isolated with varying fluoride resistance on LB agar with fluoride concentraion range between 25 mg/L to 600 mg/Lat pH 7 (Table 2). Among eight strains isolated, three strains (MB1, F and G) were showing high fluoride resistance at 500 mg/L (Figure 2), which were further explored for their role in Bioremediation of Fluoride.
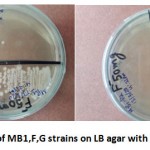 |
Figure2: Growth of MB1,F,G strains on LB agar with 50mg/100ml NaF |
Table 2: Fluoride resistant bacterial isolates grown on Luria Bertani agar with different fluoride concentrations.
| CONCENTRAION OF FLUORIDE (mg/1000ml) |
25 | 50 | 100 | 200 | 300 | 400 | 500 |
| Fluoride resistant strains | MA, MB, MB1, MB1D2, D1, E1, F, G | MA, MB, MB1, MB1D2, D1, E1, F, G | MA, MB, MB1, MB1D2, D1, E1, F, G | MA, MB1, MB1D2, D1, E1, F, G | MB1, MB1D2, D1, F, G | MB1,MB1D2, D1, F, G | MB1, F, G |
Characterization of Bacterial isolates
Three strains (MB1, F and G) showing high fluoride resistance at 500mg were characterized morphologically, physiologically and biochemically. All the three strains (MB1, F and G) were in cream colour, MB1, F were in circular shape, whereas, colonies of G strain were slightly irregular in shape. MB1 and G strains were Gram positive whereas, F strain was Gram negative. Optimum temperature for the growth of all the above three strains was 30°C. Optimum pH for the growth of MB1 and G strains was 7.5, whereas, F strain has grown optimally at pH 8. All the mentioned three organisms were protease negative. When observed Amylase activity, G strainwas Amylase positive, MB1 and F strains were Amylase negative (Table 3).
Table 3: Morphological, physiological and biochemical characteristics of the purified fluoride resistant isolates.
| Fluoride resistant strains | Colony shape | Gram’s
character |
Optimum temperature of growth | Optimum pH of growth | Amylase activity |
| MB1 | ciruclar | +ve | 30°C | 7.5 | -ve |
| F | circular | -ve | 30°C | 8 | -ve |
| G | Slightly irregular | +ve | 30°C | 7.5 | +ve |
Bioremediation of fluoride by the high fluoride resistant three strains
Bioremediation activity of high fluoride resistant three isolates (MB1, F and G) were performed on every alternate day, i.e on days 0th, 2nd, 4th, 6th, 8th and 10th. Reduced fluoride concentration was observed till 8th day, after that there was no considerable defluoridation due to the reduction in cell viability.When fluoride concentration was present at 20 mgL-1, MB1 strain showed maximum fluoride removal of 68%, whereas, F and G showed 57% and 44%fluoride removal respectively, at pH 7 and 30 °C temperature with carbon source (dextrose) 10gper 100 mL after 8 days of incubation. Results indicate that,MB1 possibly a potential fluoride resistant bacterium with high fluoride bioremediation capacity. Xu et al., 2011 reported a bacterial species with maximum of 22.1% fluoride removal.
Conclusion
In the present study, three fluoride resistant bacteria (MB1, F, G) showing high fluoride resistance (up to 500 mg/L NaF) have been purified from ground water sample of highly fluoride affected location of Nalgonda district i.e. Narketpally. The above three strains showed varying bioremediation activity, in which MB1 strain showed maximum fluoride elimination of 68%, whereas, F and G strains showed 57% and 44%fluoride degradation respectively, when fluoride concentration was present at 20 mgL-1 at 30°C temperature and pH 7 with dextrose (10 g) as carbon source per 100 mL media after incubation of 8 days. Though the MB1 strain was showing promising results in Fluoride bioremediation, further study needs to be conducted in future to achieve longevity and maintenance of bacteria through immobilization studies.
Acknowledgment
Authors feel thankful to the village people in Nalgonda district for their support during the field study.Authors feel thankful to Mr.Veera reedy, lab incharge, Water quality analysis lab, Mission Bhageeratha Section, Panagal for allowing them to analyse fluoride in samples collected from various sites of Nalgonda district ground water,in their lab.
Conflict of Interest
The authors disclosed no potential conflicts of interest, financial or otherwise.
Funding source
References
- Aguirre-Sierra A., Alonso A., Camargo J.A. Fluoride bioaccumulation and toxic effects on the survival and behavior of the endangered white-clawed cray-fish Austropotamobiuspallipes(Lereboullet). Environ.Contam.Toxicol.2013;65:244-50.
CrossRef - Ayoob S., Gupta A.K. Fluoride in drinking water: A review on the status and stress effects. Monit. Assess Crit. Rev. Environ. Sci. Technol.2006;36:433–487.
CrossRef - Banerjee G., Sengupta A., Roy T., Banerjee P., Chattopadhyaya A., Ray A. K. Isolation and Characterization of Fluoride resistant bacterial strains from Fluoride endemic areas of west Bengal, India: Assesment of their Fluoride absorption efficiency. Fl. 2016; 49(4 pt 1):429-440
- Chattopadhyay A., Podder S., Agarwal S., Bhattacharya S.Fluoride-induced Histopathology and Synthesis of stress protein in Liver and Kidney of mice.Toxicol.2011;85:327-35.
CrossRef - Choubisa S.L. Status of Fluorosis in Animals. Natl. Acad. Sci. India Sect. B Biol. Sci. 2012;82(3):331-339.
CrossRef - Chouhan S., Tuteja U., Flora S.J.S. Isolation, Identification and Characterization of Fluoride Resistant Bacteria: Possible role in bioremediation. Biochem.Microbiol.2012;48:43-50.
CrossRef - Eren E., Ozturk M., Mumcu E.F., Canatan D. Fluorosis and its Hematological effects. Ind. Health2005;21:255-8.
CrossRef - Ibrahim M., Rasheed A., Sumalatha M., Prabhakar P. Effects of fluoride contents in ground water: a review. J. Pharm. Appl.2011;2(2):128–134.
- Liao Y., Chen J., Brandt B.W., Zhu Y., Li J., Loveren C., Deng D. M. Identification and functional analysis of genome mutations in a fluoride-resistant Streptococcus mutans Plos one. 2015; 10.
CrossRef - Liao Y., Yang J., Brandt B.W., Li J., Crielaard W., Loveren C.V. Deng D.M. Genetic loci associated with Fluoride resistance in Streptococcus mutans. Microbiol. 2018.https://doi.org/10.3389/fmicb.2018.03093.
CrossRef - Maltz M., Emilson C.G. Susceptibility of oral bacteria to various fluoride salts. Dent. Res. 1982; 61: 786-790.
CrossRef - Meenakshi, MaheshwariC.Fluoride in Drinking water and its Removal. J. Hazard. Mat.2006; 137 (1): 456-463.
CrossRef - Mitsuhata C., Puteri M.M., Ohara Y., Tatsukawa N., Kozai K. Possible involvement of enolase in fluoride resistance in Streptococcus mutans. Dentl. J.2014;24:12-16.
CrossRef - Mukherjee S., Yadav V., Mondal M.,Banerjee S., Halder Charaterization of a Fluoride-resistant bacterium Acinetobacter sp. RH5 towards assessment of its water defluoridation capability. Appl. Wat. Sci.2017;7:1923-1930.
CrossRef - Podder S., Chattopadhyay A., Bhattacharya S. In vivo suppression by Fluoride of chromosome aberrations induced by mitomycin-c in mouse bone marrow cells. 2008;41(1):40-3.
- Podder S., Chattopadhyay A., Bhattacharya S., Ray M.R. Histopathology and cell cycle alteration in the spleen of mice from low and high doses of sodium fluoride. 2010;43(4):237-45.
- Susheela A.K. Dental fluorosis and its extended effect. J.Pediatr.2013;80:715-7.
CrossRef - States of the art report on the extent of Fluoride in drinking water and the resulting endemicity in India. Report by Fluorosis and Rural Development Foundation for UNICEF, New Delhi. Environ. Monit. Assess.1999;145: 1–65.
- Whitford G.M. Fluoride: Metabolism, Mechanism of action and Safety. Hyg. 1983; 57:16-8.
- WHO, “Guidelines for Drinking Water Quality (Vol. II): Health Criteria and Supporting Information, Geneva, Switzerland, 1984.
- Xu J., Song X.A., Zhang Q., Pan H., Liang Y., Fan X.W., Zhi Y. Characterization of metal removal of immobilized Bacillus strain CR-7 biomass from acqueous solutions. Hazard. Mater.2011;187: 450-458.
CrossRef - Liao Y., Brandt B.W., Li J., Crielaard W., Loveren C. V., Deng D. M., Fluoride resistance in Streptococcus mutans: a mini review. Oral Microbiol.2017;9(1):DOI: 10.1080/20002297.2017.1344509
CrossRef - Zhan X.A., Wang M., Xu Z.R., Li J.X. Toxic effects of Fluoride on Kidney function and Histological structure in young pigs. 2006;39(1):22-6.
- Zhang X., Gao X., Li C., Luo X., Wang Y. Fluoride contributes to the shaping of microbial community in high fluoride groundwater in Qiji county, Yuncheng city, china. Rep. 9, 2019; https://doi.org/10.1038/s41598-019-50914-6.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





