How to Cite | Publication History | PlumX Article Matrix
Studying the Prevalence of Mitochondrial Trnaleu Gene Mutation in Iraqi Population
Rasha Sadeq Ameen1, Mohammed Mahdi1, Dhuha Salim Namaa1, Miriam Jasim Shehab1, Suhaeer Hassan2, Reem Husam1 and Sahar Rasheed1
1Department of training and development, Forensic DNAcentre for research and training, Al-Nahrain University, Baghdad, Iraq.
2Department of clinical laboratory science, college of Pharmacy, Baghdad University, Baghdad, Iraq.
Corresponding Author E-mail: rashasadik8373@gmail.com
DOI : http://dx.doi.org/10.13005/bbra/2553
ABSTRACT: Several studies reported the role of mitochondrial gene mutations in the development of this study conducted to determine the incidence of point mutation A3243G RNALeu (UUR) in diabetes pateints within the Iraqi population and campare it with that reported in other populations. Peripheral blood were collected from 100 patients attended university of Al-Mustansiriyah / national centre for the treatment of diabetes and research. The age, gender, family history, hypertension, retinopathy, nephropathy and smoking in addition to the body mass index, are the information collected from The pateints. The DNA was extracted and by PCR-RFLP method and PCR-sequencing methods, the tRNALeu (UUR) gene screened for A3243G revealed that none of the 100 patients were found to carry the A3243G mutation in the mitochondrial tRNALeu (UUR) gene in the homoplasmic or in the heteroplasmic form. Depending on the obtained results, it can be concluded that the A3243G mutation in mitochondrial tRNALeu (UUR) is not a frequent cause of diabetes in the Iraqi population contrary to other reported populations. And further screening of an enlarged group is necessary to fully determine the prevalence of this mutation in this population.
KEYWORDS: Diabetes; Mitochondrial DNA; tRNALeu (UUR) gene; PCR- RFLP analysis
Download this article as:| Copy the following to cite this article: Ameen R. S, Mahdi M, Namaa D. S, Shehab M. J, Hassan S, Husam R, Rasheed S. Studying the Prevalence of Mitochondrial Trnaleu Gene Mutation in Iraqi Population. Biosci Biotech Res Asia 2017;14(3). |
| Copy the following to cite this URL: Ameen R. S, Mahdi M, Namaa D. S, Shehab M. J, Hassan S, Husam R, Rasheed S. Studying the Prevalence of Mitochondrial Trnaleu Gene Mutation in Iraqi Population. Biosci Biotech Res Asia 2017;14(3). Available from: https://www.biotech-asia.org/?p=27105 |
Introduction
Diabetes is a collection of diseases characterized by the presence of chronic hyperglycaemia. Maintenance of normal glucose homeostasis involves the action of a glucose sensor in thepancreatic B-cell that detects an increase in blood glucose concentration and converts that into increased secretion of insulin. Increased circulating insulin concentrations suppress hepatic glucose output and stimulate glucose uptake by muscle and adipose tissue. 1 It has been reported that some form of diabetes mellitus could be caused by mitochondrial gene abnormalities; a large deletion 2 and transition at position 3243 in the mitochondrial tRNAleu (UUR) gene of an adenine A) to guanine (G). 3 The mitochondria is an indispensable organelle for oxidative phosphorylation. Thus, it is inferred that impaired oxidative insulin secretion, thereby inducing diabetes mellitus. However, it is not clear whether decreased insulin sensitivity due to a mutant mt
DNA in the muscle is involved.4,5
Mutations in mitochondrial DNA ( mtDNA) is the Genetic factors that play an important role in the onset of type II Diabetes Mellitus, because oxidative phosphorylation in mitochondria plays an important role in insulin secretion by β-cells of the pancreas as a response to glucose and other nutrients in the body.6 Maternal Inherited Diabetes and Deafness (MIDD ) is a form of diabetes results from mutations in the MtDNA gene This form of diabetes can be diagnosed above 25 years in the form of impaired insulin secretion and is often followed by a weakening of the sense of sight and or hearing. MIDD has a very specific pattern of inheritance, through the maternal lineage without the presence of recombination of paternal line. This is because only eggs carry mtDNA when fused with sperm cells [7].An A to G substitution (guanine for adenine) at base pair 3243 in mitochondrial tRNA gene (mt3243) is commonly associated with maternally in herited diabetes and deafness, and other diseases. It is possible that cell free mitochondrial DNA exists in serum and plasmafrom these patients, and those samples might be a source of material for the detection of such mutations.8,9 Because all of that mentioned above and because of the limited study in Iraq so, This study aimed to determine the incidence of point mutation A3243G RNALeu (UUR) in diabetes pateints within the Iraqi population and campare it with that reported in other populations.
Materials and Methods
Sample Collection
Peripheral blood were collected from patients attended university of Al-Mustansiriyah/ national centre for the treatment of diabetes and research. All patients and control subjects were from all the provinces of Iraq. Patients are classified according to the following criteria (defined by the National Diabetes Data Group): age, gender, family history, hypertension, retinopathy, nephropathy and smoking in addition to the body mass index
DNA Extraction
Peripheral blood (5 mL) was collected in EDTA tubes. Nucleic acids were extracted from white blood cells by using Wizardr Genomic DNA Purification Kit (Promega, USA) according to the manufacturer’s instructions. DNA quantity and quality were assessed by Nano Drop spectrophotometer and gel electrophoresis, and stored at -20 ͦ C for further test.
Polymerase Chain Reaction
polymerase chain reaction (PCR) in an Applied Biosystems thermal cycler was performed for Genotyping the A3242G mutations, in a 25 uL reaction volume containing 0.5 uL of each primer (Canada) (Forward Primer was 5′-CGTTTGTTCAACGATTAAAG-3′ and Reverse Primer was 5′ AGCGAAGGGTTGTACTAGCC-3′ ), and 12.5 ul of go tag green master mix (Promega, USA), and 3 uL of DNA sample. A three-step PCR for A3242G was performed as described by.10 In which the first step include initial denaturation at 94’C for 5 minutes, followed by 30 cycle of 94’C for 1 minutes, 57’C for 1 minutes and 72’C for 1 minutes, with final extension at 72’C for 1 minute, The amplified products were analysed by electrophoresis on 2 % agarose gel electrophoresis. Bands were analysed and imaged by a gel documentation system.
RFLP Analysis
In order to identify the theth A-to-G mutation at nucleotide position 3243 in the tRNALeu (UUR) gene, The Fragment of PCR product with a molecular weight 422-bp in size was digested by the restriction enzyme Apa1l (Promega, USA), the digestion reaction contained 0.5ul Apall restriction enzyme, 0.5ul Bovine serum albumin, 2ul of 10X buffer, 7 μl nuclease free water and 10ul of PCR product. Followed by incubation at 37 C’ for 3 hour and electrophoreses on 3% agarose gel stained with red safe.
DNA Sequencing
To confirm the results of PCR-RFLP analysis, sequencing was performed on an 3130xl -automated DNA sequencer using Big Dye terminator kit (Applied Biosystems 313 0xl DNA Analyzer). The amplified fragment of PCR products was directly sequenced after purification by Intron Biotechnology (MEGA quick-spin Total fragment DNA purification Kit), The primers used to gain the tRNALeu (UUR) gene sequence were the same as used for the PCR amplification. The final volume of Sequencing reaction was 20 ul. Included 8 ul of Big dye terminator, 1 ul primer, 4 ul PCR product, 7 ul D.W. and the Cycling conditions was as follow : (initial denaturation at 96 ‘C for 1 minutes, followed by 25 cycle of 96 ‘C for 10 sec, 55’C for 5 sec, 60’C for 4 minutes. Sequence data analysis was performed using sequencing analysis v5.4 software. The sequencing results was analyzed using Basic Local Alignment Search independently on Ref-sequence of mitochondrial tRNA gene.
Statistical Analysis
The Statistical Analysis System- SAS (2012) was used to compare the effect of different factor in the study parameters (percentage). Chi-square test was used to compare the difference between patients and apparently healthy groups as related with genotype frequency. Odd ratio were used for determine the risk of disease.
Result and Discussion
Sample Collection
One hundred blood samples were collected diabetic and fifty from apparently healthy subjects (control). The characteristics of the patients and the control groups are summarized in (table 1). According to the age it has been shown that, The number of those with less than 40 years old in the control group was significantly (p˂0.05) higher than in patients group (18% versus 8%, respectively) and There was no significant difference between control group and patients group as related with the number within the age group 40-60 years old, while, the number of those with more than 60 years old in patients group was significantly (p˂0.05) higher than in the control group (30 % versus 18%, respectively).According to BMI 18.5 -25) there was significant difference between control group and patients group (18% versus 8%, p˂0.05), while there was no significant difference between control group and T2DM group as related with BMI (25.1 – 30). In contrast BMI 30 was in patients group significantly (p˂0.05) higher than in control group (46 % versus 38%, respectively). WHO regards underweight refereto a BMI of less than 18.5 and may indicate malnutrition while overweight refer to a BMI greater than 25 and above 30 is considered obese depending on low BMI=Mass (kg)/ Height (m)2 (WHO, 2014).the simple method to assess how much an individual’s body weight departs from what is normal or desirable is the BMI which used in a wide variety of contexts . The present analysis of BMI showed a highest ratio in the control group was overweight (BMI, 25.1 to 30) while in T2DM group was 46% overweight (BMI, 25.1 to 30) and 46% obese (BMI, more than 30).The number of males was significantly (p˂0.05) higher in the control group than in T2DM group (62% versus 50%, respectively). On the contrary, the number of females was significantly (p˂0.05) higher in T2DM group than in the control group (50% versus 38%, respectively). The percentage of family history in the control group was significantly (p˂0.01) lower than in T2DM group (24% versus 52%, respectively).There was no significant difference between the control group and T2DM group as related with smoking status. As related with hypertension, the percentage in the control group was significantly (p˂0.01) lower than in T2DM group (20% versus 56%, respectively). Also, as related with retinopathy, the percentage in the control group was significantly (p˂0.01) lower than in T2 DM group (6% versus 42%, respectively). As related with nephropathy, the percentage was significantly (p˂0.05) lower in control group than in T2DM group (2 versus 14%, respectively).
Table 1: Distribution of Apparently Healthy Subjects and type 2 Diabetes Mellitus Patients According to Some Parameters.
| Parameters | Control1
n (%) |
T2DM 2
n (%) |
p-value |
| Age (year) | |||
| ˂ 40 | 9 (18%) | 8 (8%) | 0.0355 *3 |
| 40-60 | 32 (64%) | 62 (62%) | 0.472 NS4 |
| ˃ 60 | 9 (18%) | 30 (30%) | 0.0391 * |
| Body mass index (BMI) | |||
| 18.5 – 25 | 9 (18%) | 8 (8%) | 0.0355 * |
| 25.1 – 30 | 22 (44%) | 46 (46%) | 0.472 NS |
| ˃ 30 | 19 (38%) | 46 (46%) | 0.052 * |
| Sex | |||
| Male | 31 (62%) | 50 (50%) | 0.0391 * |
| Female | 19 (38%) | 50 (50%) | 0.0391 * |
| Family history | |||
| Yes | 12 (24%) | 52 (52%) | 0.0041 **5 |
| No | 38 (76%) | 48 (48%) | 0.0125 ** |
| Smoking | |||
| Yes | 8 (16%) | 10 (10%) | 0.274 NS |
| No | 42 (84%) | 90 (90%) | 0.274 NS |
| Hypertension | |||
| Yes | 10 (20%) | 56 (56%) | 0.0015 ** |
| No | 40 (80%) | 44 (44%) | 0.0001 ** |
| Retinopathy | |||
| Yes | 3 (6%) | 42 (42%) | 0.0001 ** |
| No | 47 (94%) | 58 (58%) | 0.0001 ** |
| Nephropathy | |||
| Yes | 1 (2%) | 14 (14%) | 0.0355 * |
| No | 49 (98%) | 86 (86%) | 0.0355 * |
1 apparently healthy subject; 2 type 2 diabetes mellitus 3 p˂0.05; 4 no significant; 5 p˂0.01.
DNA Extraction
By using Wizard Genomic DNA Kit (Promega, USA),the DNA was extracted from the fresh blood samples obtained from the patients and control subjects to obtain a pure DNA for PCR amplification. The results showed that fresh blood samples yielded enough DNA concentration for PCR amplification (Fig 1), which was ranged between 50-100 ng / ul with an purity range between 1.8 – 1.9.Blood cells are targeted as sample in this study due to the sufficient number of mitochondria organelles in blood cells compared to many other cells, such as muscle cells, sperm tail cells, and hair root cells.
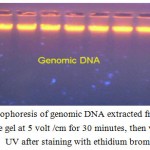 |
Figure 1: Gel electrophoresis of genomic DNA extracted from blood samples. 1% agarose gel at 5 volt /cm for 30 minutes, then visualized under UV after staining with ethidium bromide.
|
Moreover, it is relatively easy for blood sampling and has been used as a sample of previous research that has been done by 6,7,10,11 and 12 to analyze the mt DNA A3243G mutation associated with diabetes mellitus in Indonesia, Poland, Indian, Japan, and Korea respectively.
Polymerase Chain Reaction and RFLP Analysis
After DNA was extracted from T2DM patients and control subject, it has been amplified by PCR in order to obtain tRNAleu gene in vitro using the primer pair with the nucleotide sequence based on the previous study. 10 Forward Primer was covering position 3035-3054 and Reverse Primer was covering position 3437-3456. As a result of the amplication, a single band with a molecular weight 422 bp in size, as shown in ( Fig 2). It can be seen from the appearance of the band in the figure that lies parallel between the 400 bp and 500 bp. DNA from control produces a band that is located at similar position with the patients sample, while the negative control did not generate bands on gel electrophoresis indicating the absence of contaminants in the PCR process has been done. the presence of A3243G mutation in diabetic patient or not was indicated by Fragmentation of mtDNA 422 bp of tRNAleu gene by Apa1 restriction enzyme into two fragments 210 bp, and 212 bp as mentioned by.10 and 13 This happens because the A3243G mutation causes the formation of 6 nucleotides recognition site recognized by Apa1 restriction enzyme at sequence, namely GGGCCC. While for normal subjects, GAGCCC sequence is present, where concludes that there are no cut by the Apa1 restriction enzymes, because it is not the specific recognition Apa1. The Apa1 cuts, results in blunt ends to form, explains the cut right in the middle at the recognization site on the double-stranded 6 nucleotide sequence. The present of 422 bp whole band in PCR-RFLP sample results by Apa1 restriction enzyme indicates that these mutations are heteroplasmy (a mixture of mutated mtDNA and normal mtDNA in the cell). (Fig 3) reveals the results obtained after treatment of the PCR product with the restriction enzyme (Apa1), the bands do not cut by the enzyme, which refer to the incidence of diabetes is not correlated to the mitochondrial A3243G tRNAleu gene mutation.The restriction enzyme Apa1 have been used by other study to detect mtDNA A3243G mutation in DM such as 14 which studied the Incidence and prevalence of T2DM in the Egyptian populations, and detect the presence of (mt A3243G) in the serum of DM patients, and those with family history of diabetes mellitus. after polymerase chain reaction (PCR) analysis was done. The serum samples were subjected to ApaI digestion to detect any mutation. Results revealed that the presence of mt3243 allowed the 294bp product to be cleaved into 180 and 114 bp fragments and mt A3243G was detected in the serum samples of seven patients out of ten With diabetes who had positive family history of diabetes (DM). While, interestingly those with type 2 DM without family history showed positive mutation in six out of the ten patients. In the third group three out of the ten participants, showed to have a positive mutation. While none of the serum samples from healthy subjects revealed such mutations; these finding were of statistical significance (p 0.05). Therefore, they Concluded that mtDNA and associated mutations are present and detectable in the serum of patients. While in the study of Chandra and et al 6 they find out the potential of mitochondrial DNA mutation at A3243G in Indonesian type 2 diabetic Patient by use PCR-RFLP analysis. PCR products were fragment of 294 base pair (bp) and characterized by restriction enzyme ApaI. The results of their study was Heteroplasmic A3243G mutation identified in 2 Subject (0,02%) which shown by 3 electrophoretic bands, 2 restriction products of APAI, i.e a 182 bp fragment and a 112 bp fragment; also a full fragment 294 bp, approved that PCR-RFLP technique can be used to identify heteroplasmic A3243G mutation in a mt DNAtRNAleu gene. Dorraj et al 15 mentioned that the A3243G tRNALeu mutation was not detected in Iranian patients with type 2 diabetes after Digestion with ApaI lee et al., 1997 12 studied (tRNA)Leu (UUR) 3243 and tRNA Lys 8344 Mutations in Diabetes patients from Korea . after digestion the PCR product with endonucleases ApaI and if theremt DNA mutation at the 3243 position the fragment cleaved in to two fragments, Also in the study of 16 and 13 they use the Apa1 to detect the A3243G tRNAleu gene mutation in Nigereia and Pakistani population respectively.
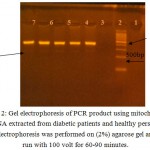 |
Figure 2: Gel electrophoresis of PCR product using mitochondrial DNA extracted from diabetic patients and healthy person , Electrophoresis was performed on (2%) agarose gel and run with 100 volt for 60-90 minutes.
|
The lanes: 1: hyperladder IV DNA marker (100bp). 2:negative control. 3,4,5: DNA from diabetics patients 6,7: DNA from control
 |
Figure 3: Screening of the A3243G mutation by PCR-RFLP. PCR fragment of 422 bp was digested with ApaI.
|
The wild type PCR product does not contain the ApaI restriction site but PCR product with A3243G is expected to cleave into two fragments of 210 and 212 bp.Hence, Lanes 2 to 11 is a representation of restriction assay in patient’s samples that resulted in an uncut fragment of 422 bp while Lane 1 is the 100 bp DNA ladder (molecular marker).
The A3243G tRNAleu Sequence Analysis
Sequencing the PCR product (422bp) was done to proof the results of PCR-RFLP analysis, in which DNA of patients and control subject were screened for mutation by comparing their sequences with the annotated human mitochondrial genome database in order to identify the A3243G mutation. The identified results indicated that there is no correlation between the incidence of diabetes and A3243G tRNALeu(UUR) gene m utation in Iraqi patients. It is may be the reasons which led to non detection of the mutation belong to the blood sample which is used as the source of the mtDNA. different samples like blood, hair follicles, buccal epithelial cells and muscle biopsy have been used to determine the presence of A3243G mutation by other study such as 17,18 and they shown that the proportions of A3243G mutant in diabetic subjects were noted to be the highest in muscle tissue followed by hair follicles and lowest in blood cells. Interestingly, the proportions of this mutation in all the tissues were found to decline with increase in age18
It is also noted that the obtained results were unlike that from other Clinical studies of mtDNA point mutation A3243G in DM patients that conducted in various countries. In which mutations are found with different presentations, such as in Taiwan it is found in 0.15% of the entire population of patients with Diabetes Mellitus,19 whereas in Poland, number of patients with A3243G mutations are not known for sure. 7 Point mutations in the mt DNA A3243G is also found in Japan 2.9%,10 in England 0.75% 20 and in Croatia 10%.21 While in Korea 22.3% patient with mitochondrial disease had point mutation on the A3243G mtDNA.22 In Spain it has been found in 18% of children patients had three heteroplasmy mutations including A3243G.23 While the A3243G mutation frequency was 2. 22% in the investigated patients of hungary,24 in Indian 7.8% 11 and in Indonesian patients it was identified in 2 Subject (0,02%).6 This mutation not only affects the synthesis of tRNAleu but also interfere with the binding mechanism of transcription termination factor that may lead to disruption of the synthesis of mitochondrial proteins. 25 According to the study done by 26,15,13 and 16 were unable to detect the A3243G mutation in diabetic patients, in Iranian, Pakistani and in the Nigerian population respectively, in which they closely related to the results obtained in this study And unlike the result obtained in the studyof 27 who revealed that both the mother and fetus carried the MELAS-specific A3243G mutation in pregnant Taiwanese women. Also in they study, mentioned that the mtDNA level of the amniotic fluid did not significantly differ from that of the postnatal peripheral blood and hair follicles. And concluded that MELAS syndrome may help in identifying mitochondrial mutations during pregnancy.
Conclusions
It has demonstrated from the results that has been obtained, that there is no correlation between the mitochondrial DNA mutation and the incidence of diabetes in Iraqi patients in comparison with other reported investigation. And further study are required with enlarge number of subjected patient for screening, and with different type of sample like hair, tissue and muscle to be ensure that This mutation is not significance in diabetes mellitus pathogenesis in Iraqi populations.
Acknowledgements
Authors would like to thank the university of Al-Mustansiriyah/ national centre for the treatment of diabetes and research, also, Al-Nahrain University, Forensic DNA centre for research and training for the financialsupport to this study.
Fund
No fund was received for doing this study.
References
- Maassen J. A., Hart L. M., Essen E., Heine R. J., Nijpels G., Tafrechi R. S. J., Raap A. K., Janssen G. M. C., and Lemkes H. H. P. Mitochondrial Diabetes Molecular Mechanisms and Clinical Presentation. DIABETES. 2004;53(1):103-109.
CrossRef - Ballinger S. W., Shoffner J. M., Hedaya E. V., et al. Maternally transmitted diabetesand deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat Genet. 1992;1:ll.
CrossRef - van den., Ouweland J. M., Lemkes H. H., Ruitenbeek W., et al. Mutation inmitochondrialtRNAleu(UUR)gene in a large pedigree with maternallytransmitted type II diabetes mellitus and deafness. Nat Genet. 1992;1:368.
CrossRef - Shigemoto M., Yasunao Y., Yuji Y., Tatsuya H., Junko S., Gen I., Motozumi O., Hisato J., Kinsuke T., Toshikazu Y., Toshihiro Y., Mariko O., Satoshi T., Hideshi K and Kazuwa N. Clinical Manifestations due to a Point Mutation of the Mitochondrial tRNAleu(UUR) Gene in Five Families with Diabetes Mellitus. Internal Medicine. 1998;37(3):265-272.
CrossRef - Wolf N. I and Smeitink J. A. M. Mitochondrial disorders. Neurology. 2002;59:1402–5.
CrossRef - Chandra R. A., Ajeng D. S and Iman P. M . Restriction Enzymes ApaI Analysis to Find A3243G Mutation in Indonesia Diabetes Mellitus Type II Patients. Journal of Medical and Bioengineering. 2015;4( 6):492-496.
CrossRef - Malecki M., Klupa T., Wanic K., Frey J., Cyganek K and Sieradzki J. Search for mitochondrial A3243G tRNAleu mutation in Polish patients with type 2 diabetes mellitus. Med Sci Monit. 2001;7(2):246-250.
- Sheng Z., Maggie C. Y., Dennis Y. M., Juliana C. N., Philip J. J. Presence of mitochondrial tRNALeu(UUR) A to G 3243 mutation in DNA extracted from serum and plasma of patients with type 2 diabetes mellitus. J Clin Pathol. 2000;53:466-469.
CrossRef - Padma V. V ., Anitha S., Santhini E., Pradeepa D., Tresa D., Ganesan P., Ishwarya P., Balamurugan R. Mitochondrial andnuclear gene mutations in the type 2 diabetes patients of Coimbatore population. Mol. Cell Biochem. 2010;345(1-2):223-9.
CrossRef - Ohkubo K., Yamano A., Nagashima M., Mori Y., Anzai K., Akehi Y., Nomiyama R., Asano T., Urae A and Ono J. Mitochondrial gene mutations in the tRNALeu(UUR) region and diabetes prevalence and clinical phenotypes in Japan. ClinChem (Mol Diagnost Genet). 2001;47(9):1641–8.
- Khan I. A., Shaik A. N., Pasupuleti N., Chava S., Jahan P., Hasan Q. P. Screening of mitochondrial mutations and insertion–deletion polymorphism in gestational diabetes mellitus in the Asian Indian population. Saudi Journal of Biological Sciences. 2015;22:243–248.
CrossRef - Lee C. H., Song D. Y., Li R . H., Park O. J., Suh C. H., Lee E., Lim S., Kim K and Huh K. Mitochondrial Gene Transfer Ribonucleic Acid (tRNA)Leu(UUR) 3243 and tRNALys 8344 Mutations and Diabetes Mellitus in Korea. Journal of Clinical Endocrinology and Metabolism. 1997;82( 2):372-374.
CrossRef - Naveed A. K., Wahid M and Naveed A. Mitochondrial tRNALeu(UUR) gene mutation and maternally inherited diabetes mellitus in Pakistani population. International Journal of Diabetes Mellitus. 2009;1:11–15.
CrossRef - EI-Gaaly S. A., Radwan H. H., ElSherbeny A. A and Eldin M. A. S. The Genetic Mutation in patients with Type 2 Diabetes Mellitus in Adult. Life Science Journal. 2014;11(10s):683- 687.
- Dorraj G., Houshmand M., Larijani B., Majd A., Hosseini B., Montazeri M., Panahi M. S . Lack of association of mitochondrial A3243G tRNALeu mutation in Iranian patients with type 2 diabetes. Iranian journal of biotechnology. 2005;3(4):243- 248.
- Ameh J., Godwin I., Obi I., Puepet F., Aminu B and Suleiman T. The search for mitochondrial tRNALeu (UUR) A3243G mutation among type 2 diabetes mellitus patients in the Nigerian population African Journal of Biotechnology. 2011;10(62):13383-13389.
- Kari M., Jukka S. M., Seija U., Anne M. R., Pasi I. S., Mikko K. Epidemiology of A3243G, the mutation for Mitochondrial Encephalomyopathy, Lactic Acidosis and Strokelike Episodes Prevalence of the Mutation in an Adult Population. Am. J. Hum. Genet. 1998;63:447-454.
CrossRef - Chia-Wei L., Chin-Chang H., Yau-Huei W. Molecular analysis of diabetes mellitus associated A3243G mitochondrial DNA mutation in Taiwanese cases. Diabetes Res. Pract. 2001;54(2):S39-S43.
- Liou C. W., Huang C and Wei Y. H. Molecular analysis of Diabetes mellitus associated A3243G mitochondrial DNA mutation in Taiwan cases. Diabetes Res. and Clinical Practice. 2001;54(2):39-43.
CrossRef - Owen K.,Sride A., Ellard S and Hattersley A . T. Etiological investigation of diabetes in young adults presenting with apparent type 2 diabetes. Diabetes Care. 2003;26(7):2088-2093.
CrossRef - Kleiner I. M., Medvidoic E. P. P.,et al. A pilot study of mitochondrial DNA point mutation A3243G in a sample of Croatian patient having type-2 diabetes mellitus associated with maternal heritance. Acta Diabetol. 2004;41:179-184.
CrossRef - Chae J., Hwang H., Lim C., et al. Cllinical features of A3243G mitochondrial tRNA mutation. Brain & Dev. 2004;26:459-462.
CrossRef - Uusimaa J., Finnila S., Remes A. M., et al. Molecular epidemiology of childhood mitochondrial encephalomyophathies in finnish population sequence analysis of entire mtDNA of 17 children reveals heteroplasmic mutation in tRNA Arg. tRNAGlu, and tRNALeu (UUR) genes. Pediatrics. 2004;112( 2):443-450.
CrossRef - Gal A., Komlosi K., Maasz A., Pentelenyi K., Remenyi V., Ovary C., Valikovics A., Dioszeghy P., Bereczki D., Melegh B., Molnár M. Analysis of mt DNA A3243G mutation frequency in Hungary. Eur. J. Med. 2010;5(3): 322-328. DOI: 10.2478/s11536-009-0118-2.
CrossRef - Kadowaki T.,Kadowaki H., Mori Y.,et al. A subtype of diabetes mellitus a mutation of mitochondrial DNA. NEJM. 1994;33:962-968.
CrossRef - Chen Y., Liao W. X., Roy A. C., Loganath A., Ng S. C. Mitochondrial gene mutations in gestational diabetes mellitus. Res. Clin. Pract. 2000;48:29–35.
CrossRef - Chou Y. J., Ou C. Y., Hsu T. Y., Liou C. W., Lee C. F., Tso D. J., Wei Y. H. Prenatal diagnosis of a fetus harboring an intermediate load of the A3243G mtDNA mutation in a maternal carrier diagnosed with MELAS syndrome. Diagn. 2004;24:367–370.
CrossRef - World Health Organisation. Global status report on non communicable diseases 2010. Geneva: WHO, 2011. Available at: http://www.who.int/nmh/publications/ncd_report_full_en.pdf. Accessed in December. 2013.

This work is licensed under a Creative Commons Attribution 4.0 International License.





