How to Cite | Publication History | PlumX Article Matrix
Stability and Mobility of Lid Lipmnk in Acetonitrile by Molecular Dynamics Simulations Approach
Dian Herasari1,2, Rukman Hertadi1, Fida M. Warganegara1 and Akhmaloka1,3
1Biochemistry Research Group, Faculty of Mathematics and Natural Science, Institut Teknologi Bandung, Indonesia.
2Department of Chemistry, Faculty of Mathematics and Natural Science, Universitas Lampung, Indonesia.
3Department of Chemistry, Faculty of Science and Computer, Universitas Pertamina, Indonesia.
Corresponding Author E-mail: loka@chem.itb.ac.id
DOI : http://dx.doi.org/10.13005/bbra/2632
ABSTRACT: Manuk lipase (lipMNK) from the thermophilic bacterium Geobacillus sp is a double lid lipase containing short and long lid segments. A few studies demonstrated that catalytic action of lipase involved the movement of lid segments from closed to open conformation upon the substrate binding. One factor that affects conformational dynamics of the lid segments is solvent polarity. The presence of acetonitrile in certain concentration has showed to enhance lipase activity. In this study, the effect of acetonitrile to the stability and activity of lipMNK was studied at the atomic level by molecular dynamics (MD) simulation. MD was carried out by NPT ensemble at 358 K for 100 nano seconds in various ratio of acetonitrile:water solvent mixtures. The results showed that the conformation of lipMNK was stable up to 70%. However, the effect of lid movement was significantly observed since the concentration at 20% acetonitrile. Detailed molecular analysis at this acetonitrile concentration revealed that the two lids moved in different modes upon opening and closing movement. In the opening movement, the two lids appeared to move in almost simultaneously, while during the closing movement, it was observed sequentially, started by short segment followed by long segment lid.
KEYWORDS: lipMNK; lid; Molecular Dynamics
Download this article as:| Copy the following to cite this article: Herasari D, Hertadi R, Warganegara F. M, Akhmaloka A. Stability and Mobility of Lid Lipmnk in Acetonitrile by Molecular Dynamics Simulations Approach. Biosci Biotech Res Asia 2018;15(2). |
| Copy the following to cite this URL: Herasari D, Hertadi R, Warganegara F. M, Akhmaloka A. Stability and Mobility of Lid Lipmnk in Acetonitrile by Molecular Dynamics Simulations Approach. Biosci Biotech Res Asia 2018;15(2). Available from: https://www.biotech-asia.org/?p=29922 |
Introduction
Lipase is hydrolase class of enzymes that play role in the hydrolyzing of triacylglicerol. Lipase has been widely applied in many industries, including in the food, detergent, waste processing, textile, pharmaceutical and pulp or paper industries.1-4 Due to its wide and varied applications, industrial lipases must able to effectively work in various industrial conditions that may involve on of high temperature, some organic solvents and others. Therefore, multistable lipases are highly required by some industries to enhance the process and reduce the production cost.
One approach to obtain multistable lipases are by exploring extremophilic microorganisms as the enzyme sources. Thermophilic bacteria are the best source to obtain thermostable enzymes. In the previous study, we have cloned lipase gene from the thermophilic bacteria Geobacillus sp indigenous of Manuk Crater, West Java, Indonesia.5 The recombinant lipase expressed from the gene was named as lipMNK. The gene contains 416 amino acids residues comprising of 28 residues of the signal peptide and 388 structural residues. The enzyme has two lids in its catalytic domain with different length. The long lid, which is located at residues of 175 to 195 from N-terminal, namely lid A, containing amino acid sequence of DETDRFFDLQKAVLEAAAVAS, while for short lid, located at 221 to 230 residues from N-terminal with the sequence of FDHYFERLKR, namely lid B.6 The partially purified enzyme showed an optimum activity at pH 7 and 85oC.7
Besides thermal activation, several studies found that modifying solvent polarity by addition of certain organic solvent also improve lipase activity.8 Therefore, organic solvent stable lipase is also interesting for industrial applications, particularly involving organic molecule as its substrate that merely dissolved in non-polar solvent. One of the organic solvents that significantly increase lipase activity is acetonitrile.9 It is miscible polar organic solvent taken a role as water’s co-solvent for lipase. Several studies on the effect of acetonitrile on lipase activity suggest different conclusions. The activity of lipase from Geobacillus sp. strain ARM was inhibited when acetonitrile was added to the enzyme solution.10 Other studies found that the presence of acetonitrile enhanced lipase activity, such as lipase from Staphylococcus saprophyticus M3611 and from Geobacillus thermoleovorans PPD2.12 Other research suggested that acetonitrile is a potential solvent for transesterification activity of lipase.13 Therefore, it is interesting to study how the action of acetonitrile to affect the stability and activity at the molecular level of the enzyme reaction.
In this study, molecular dynamics (MD) simulation was used to observed the effect of acetonitrile on the stability and activity of lipMNK. The enzyme stability was studied by analyzing conformational change during the movement of lid segments from closed to open conformation. It is carried out by comparing the structure of lipase in various acetonitrile concentrations compared to that in the water. While for the enzyme activity, evaluation was based on how acetonitrile induced the movement of the two-lid of lipMNK in both opening and closing process. MD simulation provide molecular insight into the effect of acetonitrile to the stability and activity of lipMNK.
Materials and Methods
LipMNK Structure Model
The lipMNK structure model was prepared by structural prediction with comparative modeling using the Swissmodel program.14 Two initial structures of lip MNK at closed-lid and open-lid conformation, were constructed through the program. Two enzyme structures were used as template. Lipase L1 of B. stearothermophilus (PDB 1KU0) with 98% identity was used as closed lid template of lipMNK, the generated structure manely as c-lipMNK. While for the open lid conformation, BTL2 lipase from B. thermocatenulatus (PDB 2W22) was used. As template and generated structure was named as o-lipMNK. Comparative modeling was carried out in three main stages: alignment of amino acid sequence of lipMNK with the template structure, 3D structure prediction and structural validation.
Molecular Dynamics
A computer equipped with NVIDIA GeForce GTX 570 by CUDA® with an Intel i7 8 core processor was used for simulation. The hardware was also equipped with Ubuntu 12.04 LTS operating system and some other software. Solvation of lipMNK by acetonitrile:water mixture was performed by using Packmol program 15. All of the simulations were performed using AMBER 12 software with AMBER ff03.r1 force field. The system was minimized using SANDER program by 2,000 cycles comprising of 500 cycles with Steepest Descent Algorithm and then followed by 1.500 cycle with Conjugate Gradient Algorithm by applying restraint force to the protein backbone. The minimization procedure was repeated by gradually decreased restraint force until no restraint condition applied. Furthermore the temperature of the system was gradually heated up to 358 K in NVT ensemble and equilibrated in NPT ensemble at 358 K with a constant pressure of 1 bar. In these stages, non-bonding interactions were confined within a cut off at 12 Å. The result of equilibrated system was then used in the production run using the same simulation parameters. All production run were performed for 100 ns and saved every 1 ps resulting 100,000 structures stored in the simulation trajectory.
Analysis
The trajectory of all simulations was evaluated by calculating root mean square deviations (RMSD) for the lid backbone using both structure (close and open state) as reference and analyzed with visual molecular dynamics (VMD) program version 1.9.1.16 Root mean square deviation (RMSD) was measured for position of particle with respect to reference position over time.17 RMSD is calculated by the following formula:
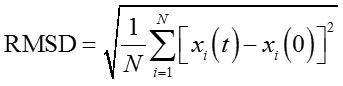
With N is the number of Cα atoms present in the protein, xi (0) is the reference position of each Cα atoms, and xi (t) is the position of each Cα atoms in determined time t.
Results and Discussions
Effect of Acetonitrile on lipMNK Stability
The conformational change of the lid from closed to open, directing lipase to its catalytic function, was known to occur at the interface between lipid and water layers18 (Jaeger et al., 1999). In this study, the lipid-water interface was created by mixing acetonitrile and water as the solvent for lipMNK.
The effect of acetonitrile to lipMNK stability was studied by performing MD simulations in different acetonitrile concentrations from 0% to 100%. The structure of closed lid conformation of lipMNK (c-lipMNK) was used in this study. All MD simulations were run for 100 ns. The structure of c-lipMNK after 100 ns of simulation in water (0% of acetonitrile) was used as the reference structure to evaluate conformational change induced by the addition of acetonitrile. The result of RMSD calculations for the structure of lipase after 100 ns simulations in variation of acetonitrile concentrations revealed various deviation compare to that the structure in water (Fig 1), which is ranging from 3 to 6 Å. RMSD value of c-lipMNK in acetonitrile concentration of 20 – 70% were below 4 Å however significant deviation was appeared at acetonitrile greater than 70%, with almost 6 Å.
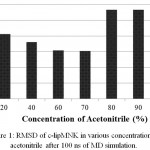 |
Figure 1: RMSD of c-lipMNK in various concentration of acetonitrile after 100 ns of MD simulation.
|
RMSD was calculated using c-lipMNK structure in 0% acetonitrile at 100 ns of MD simulation as reference. Å o = Angstrom.
Further analysis to c-lipMNK structure revealed the source of deviation in RMSD value (Fig 2). At acetonitrile concentration within 20%-70%, there were no structural denaturation observed in c-lipMNK. The deviation was only appeared due to the lid movement induced by acetonitrile. However, at 80%-100% acetonitrile concentrations, denaturation at certain region in c-lipMNK was observed. At 80% and 90% acetonitrile, structure of c-lipMNK segment at 196-220 residues were unfolded. Moreover, at 100% of acetonitrile the residues of 300 -310 was also unfolded. Both regions were random coil loop in the native structure of c-lipMNK, hence they were relatively easier to be denatured by acetonitrile. The other parts of the proteins, however, were relatively unaffected by acetonitrile suggesting that the regions were stable and acetonitrile may be used as the solvent for c-lipMNK.
 |
Figure 2: Superimpose between lipMNK structure after 100 ns MD simulation in various acetonitrile concentrations (red) and structure of lipMNK at 0% acetonitrile (blue).
|
The unfolded segment due to the presence of acetonitrile was pointed by arrow. All protein structures were generated in cartoon model by VMD.
The Effect of Acetonitrile to the lid Opening Movement of lipMNK
Several studies found that lipase activity is triggered by the lid movement induced by the substrate and the product of reaction.19 The lid movement from closed to open conformation is occurred on the substrate binding, while the movement of lid from open to closed conformation is stimulated when the product is released. Such dynamics lid movement may be modelled by changing the polarity of solvent. Previous study based on molecular dynamics simulations showed that non-polar organic solvents induced lid opening movement, while the reverse was stimulated in water solvent.20 In this study, acetonitrile was used to understand the opening lid process while water solvent was used to observe the process of closing lid.
c-lipMNK structure was used as initial structure for opening process movement while o-lipMNK structure was used as final opening conformation. The effect of acetonitrile concentrations for opening lid process were varied (Fig 3). At the acetonitrile concentrations of 20, 40, 80, 90 and 100% appeared the movement of lid, however, at concentrations of 0, 60 and 70% there were almost no detectable movement of lid. 0% acetonitrile was used as negative control for opening lid movement since no organic molecules substitute the role of lipid substrate plays role to induce the opening of lid. Among those acetonitrile concentrations that triggered opening lid, none of them was successfully forcing the lid to reach fully open conformation compared to that to o-lipMNK structure after 100 ns of simulation. Therefore, the effect of acetonitrile is not as strong as the other reported organic solvent, such as toluene, in inducing the lid opening movement,21 which is almost fully open lid.
 |
Figure 3: Snapshot of lid opening movement in various acetonitrile concentrations.
|
Initial lid position was presented in red, while the lid position after 100 ns was depicted in yellow. The other part of the protein was presented as gray and blue ribbons for the initial state and after 100 ns simulation. All structures were generated in new cartoon model by VMD.
It was surprising that at 60 and 70% acetonitrile, the lid movement were almost undetectable. Further analysis showed that at both concentrations, acetonitrile molecules did not fully solvate the lid region of lipMNK (Fig 4). There were only very small number of acetonitrile molecules detected within 5 Å from the lid region. As result, there was no interface induced for opening lid movement.
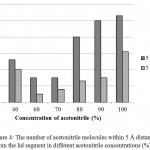 |
Figure 4: The number of acetonitrile molecules within 5 Å distance from the lid segment in different acetonitrile concentrations (%).
|
Among acetonitrile concentrations that triggered the opening lid movement, only at concentration of 20 and 40% retained the native structure of lipMNK. At acetonitrile concentrations of 80, 90 and 100%, the lid movement appeared, however there were also found unfolded conformation on the outside region of lid (Fig 3). The result suggested that only acetonitrile concentration of 20-40% were safely used to induce the opening lid movement. Other research suggests that interaction of enzymes with organic molecules may cause inactivation.22 At the interface between organic and water molecules, unfavorable condition may be occurred since enzyme molecules that adsorbed into such interface may be denatured, which is known as the interfacial inactivation.20 So that, 20% acetonitrile is considered as the safest concentration to accelerate opening movement of the lid.
lipMNK contains two lid segments, i.e. lid A (long segment) and lid B (short segment). In order to understand the pattern of opening lid movement further analysis was carried out on the simulation at 20% acetonitrile. The result showed that the opening movement of lid was initially observed at 20 ns, after that it was almost steady up to 40 ns, while at 60 ns the opening lid continuously increased (Fig 5). At the above time interval, both lid A and lid B were likely moved in simultaneously. Further evaluate by measuring the inter-lid angle based on the angle Ser195, Asp175 and Phe221 showed that in the initial processs, inter-lid angle was about 30oC and within 40 ns the angle increased up to 60oC, however, it was monotonously decreasing up to 10oC at 73 ns, later on from 80 to 100 ns, the angle was back to initial angle at 30oC. These data suggest that lid A and lid B moved independently toward the open conformation.
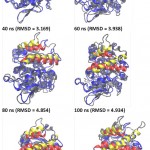 |
Figure 5: Snapshot of lid movement for every 20 ns simulation.
|
The initial and final conformation of lid in every 20 ns of the simulation were represented by red and yellow ribbon. The other part of the protein were presented as gray and blue ribbons for the initial conformation and every 20 ns of simulation. All structures were generated in new cartoon model by VMD. Circle was point out of lids; ns = nano second.
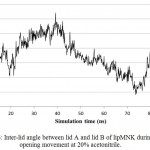 |
Figure 6: Inter-lid angle between lid A and lid B of lipMNK during lid opening movement at 20% acetonitrile.
|
The inter-lid angle was measured based on the angle formed by Cα position of Ser19, Asp175, and Phe221.
The Influence of Acetonitrile on Closing Movement at lipMNK lid
The catalytic process of lipase is not only influenced by opening but also by closing movement of the lid. The closing movement was observed by using o-lipMNK structure as the initial and c-lipMNK structure as the final closed conformation. To study the effect of acetonitrile to lid closing movement, the simulations were carried out in 0 and 20% of acetonitrile. For these simulation the distance of lid A and lid B to the final closed conformation were measured. The result showed that the distance of each lid to the final conformation was relatively similar (Table 1). This suggested that 20% of acetonitrile did not significantly influence on the closing movement of lid.
Table 1: The distance between Cα of Ser195 (lid A), Cα Phe221 (lid B) and the correspondence residues at the final closed conformation. Å = Angstrom.
| Acetonitrile concentration (%) | Lid A (Å) | Lid B (Å) |
| 0 | 13.20 | 7.87 |
| 20 | 13.14 | 7.92 |
Further analysis to understand the closing movement pattern, the RMSD in 20% acetonitrile were measured for the period of simulation time. The result showed that there was closing movement of the lid at 20 ns, however, up to 80 ns there was no other significant movement detected. Furthermore, at 100 ns both lids were observed significantly moved toward close conformation (Fig 7). The measurement of the distance between representative residue in each lid and selected residue at non lid region (Ser 195, Phe 221, and Ala289 representative of lid A, lid B, and non lid region respectively) showed that the closing movement was devided into two phases. The first phase, lid B (short lid) gradually approached lid A up to 90 ns simulation period. After the two lids close each other, the second phase was started by moving both lids toward closed conformation (Fig 8). This pattern was similar to that of Psedomonas aeruginosa lipase (PAL) 23. PAL is known as double lids lipase which is similar to lipMNK. The short lid of PAL played role to induce the long lid moved toward the close conformation.
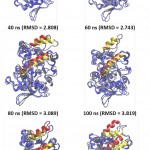 |
Figure 7: The snapshot of lid closing movement every 20-ns time interval.
|
Red and yellow helices represent lids at the initial (open) and final conformation at each time interval, respectively. The other parts of protein conformation were represened as blue and gray for the respective initial and at the time interval.
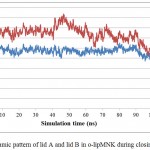 |
Figure 8: Dynamic pattern of lid A and lid B in o-lipMNK during closing movement.
|
Blue and red lines represent the respective distance covered by Ser195 (lid A) and Phe221 (lid B) relative to Ala289 (non lid region).
Conclusion
Addition of acetonitrile still maintains the native structure of lipMNK up to the concentration of 70%. However, the dynamics of lid toward opened and closed conformation that retain native structure was occurred at minimum concentration of acetonitrile at 20%. The pattern of opening and closing lid movement are unlikely not in the same manner.
Acknowledgment
The authors are grateful for the support of P3MI Research Project of ITB, Ministry of Research Technology and Higher Education of Republic of Indonesia, to make the research possible to be carried out.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with the respect to the research, authorship, and/or publication of this article.
References
- Bjorkling F, Godtfredsen S.E, Kirk O. The Future Impact of Industrial Lipases. Review. Trends Biotechnol. 1991; 9: 360-363.
CrossRef - Jaeger K.E, dan Reetz M.T. Microbial Lipases from Versatile Tools for Biotechnology. Trends Biotechnol. 1998; 16: 396-403.
CrossRef - Sharma R, Chisti Y, Banarjee U.C. Production, Purification, Characterization and Application of Lipases. Biotechnology Advances. 2001; 19: 627-662
CrossRef - Gupta R, Gupta N, Rathi P. Bacterial lipases: an overview of production purification and biochemical properties, Applied Microbial Biotechnology. 2004; 64: 763-781.
CrossRef - Widhiastuti M.P, Febriani Yohandini H, Moeis M.R, Warganegara F.M, Akhmaloka. Characterization and identification of thermostable alkaline lipase producing bacteria from hot spring around West Java, Journal of Pure and Applied Microbiology. 2009; 3(1): 2740
- Widhiastuty M.P, Moeis M.R, Akhmaloka. Cloning, homological analysis and expression of lipase gene from Manuk Hot Spring Isolate, International Journal of Integrative Biology. 2011; 11 (1): 8-13.
- Briliantoro R, Zidny R, Widhiastuty M.P, Madayanti F, Akhmaloka. Hydrolitic and Transesterification Activities of Thermostable Lipase ITB1. Biosciences Biotechnology Research Asia. 2015; 12(1): 01-06.
CrossRef - Sharma S and Kanwar S.S. Organic Solvent Tolerant Lipases and Applications. The Scientific World Journal. 2014; 15 pages.
- Lee D, Koh Y, Kim K, Kim B, Choi H, Kim D, Suhartono M.T, Pyun Y. Isolation and characterization of a thermofilic lipase from Bacillus thermoleovorans ID-1, FEMS Microbiology Letters. 1999; 19: 393-400.
CrossRef - Ebrahimpour A, Rahman R.N.Z.R, Basri M, Salleh A.B. High level expression and characterization of a novel thermostable, organic solvent tolerant, 1,3-regioselective lipase from Geobacillus sp. strain ARM. Bioresource Technology. 2011; 102: 6972 – 6981.
CrossRef - Fang Y, Lu Z and Lv F. A newly isolated organic solvent tolerant Staphylococcus saprophyticus M36 produced organic solvent-stable lipase. Current Microbiology. 2006; 53: 510.
CrossRef - Permana A.H, Warganegara F.M, Wahyuningrum D, Widhiastuty M.P, Akhmaloka. Heterologous Expression and Characterization of Thermostable Lipase. Bioscience Biothecnology Research Asia. 2017; 14(3): 1081-1088.
CrossRef - Pohlnlein M, Finkbeiner T, Syldatk C and Hausmann R. Development of microtiter plate-based assay for the detection of lipas-catalyzed transesterifications in organic solvents. Biotechnol Lett. 2015; 37: 705-710.
CrossRef - Martínez L1, Andrade R, Birgin EG, Martínez JM. PACKMOL: a package for building initial configurations for molecular dynamics simulations. Journal of Computational Chemistry. 2009; 13: 2157-2160.
CrossRef - Humphrey W, Dalke A and Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996; 14(1): 33-38, 27-28.
- Jaeger K.E, Djikstra B.W, Reetz M.T. Bacterial Biocatalysts: Molecular Biology, Three-Dimensional Structures, and Biotechnological Applications of Lipases. Annual Review of Microbiology. 1999; 53: 315-351.
CrossRef - Cajal Y, Svendsen A, Girona V, Patkar S.A, Alsina M.A, Interfacial Control of Lid Opening in Thermomyces lanuginose Lipase. Biochemistry. 2000; 39: 413-423.
CrossRef - Kumar A, Dhar K, Kanwar S.S, Arora P.K. Lipase catalysis in organic solvents: advantages and applications. Biological Procedures Online. 2016; 18:2
CrossRef - Ghatorae A.S, Guerra M.J, Bell G, Halling P.J, Immiscible organic solvent inactivation of urease, chymotrypsinm lipase, and ribonuclease separation of dissolved solvent and interfacial effect, Biotechnol Bioeng. 1994; 44(11): 1355-1361
CrossRef - Cherukuvada S.L, Seshasayee A.S.N, Raghunathan K, Anishetty S, Pennathur G, Evidence of a Double-Lid Movement in Pseudomonas aeruginosa Lipase: Insights from Molecular Dynamics Simulations. PLoS Computational Biology. 2005.
CrossRef

This work is licensed under a Creative Commons Attribution 4.0 International License.





