How to Cite | Publication History | PlumX Article Matrix
Hema Gunti*1 , Vyshali Venkatappa Maruthiramaih2
, Vyshali Venkatappa Maruthiramaih2 and Tippeswamy Boreddy Shivanandappa3
and Tippeswamy Boreddy Shivanandappa3 
1Department of Biotechnology, Maharani’s Science College for Women, Bangalore, Karnataka, Indai.
2Department of Biotechnology, BMS College for Women, Bangalore, Karnataka, India.
3Department of Biomedical Science, College of Pharmacy, Shaqura University, Kingdom of Saudi Arabia
Corresponding Author E-mail: hema_h_t@yahoo.co.in
DOI : http://dx.doi.org/10.13005/bbra/2875
ABSTRACT:
Untreated effluents from the textile industry affect aquatic life irreversibly. Synthetic dyes not only change the color of water resources but also make them hazardous.The main objective of the study was to evaluate the decolorizing potential of a new isolate of Bacillus subtilis from soil samples contaminated with industrial effluent in and around textile industrial area in South Karnataka. This isolate of Bacillus subtilis has high decolorizing potential and took only 24 hrs for complete decolorization of acid orange-10 azo dye at 200ppm. Different parameters like temperature, pH, aeration, dye concentration and inoculum size were optimized for complete decolorization of Acid orange-10 azo dye by this isolate of Bacillus subtilis. The dye was completely decolorized at 400C within 24 hrs and it was capable of decolorizing 700 ppm dye in 72 hrs. Optimum pH was found to be 8.5 and maximum decolorization was achieved under static conditions. As the inoculum size increased, the time taken for complete decolorization of Acid orange-10 dye was decreased from 36 hrs at 1% to 16 hrs at 10% of inoculum size. The new isolate decolorizes 100 ppm of dye completely (i.e.100%) within 12hrs of incubation. The time taken for the complete decolorization increased with increase in the concentration of Acid orange-10 azo dye. In conclusion, the new isolate of Bacillus subtilis from soil samples contaminated with textile industrial effluent was found to be a potential candidate for decolorization of Acid orange-10 azo dye in textile effluents.
KEYWORDS: Acid Orange-10, Azo Dye; Bacillus Subtilis; Decolorization; Optimization Of Parameters; Textile Effluent
Download this article as:| Copy the following to cite this article: Gunti H, Maruthiramaih V. V, Shivanandappa T. B. Bio-degradation of Azo Dye Acid Orange-10 by a New Isolate of Bacillus subtilis Isolated from Soil Sample around Textile Industry in South Karnataka.Biosci Biotech Res Asia 2020;17(4). |
| Copy the following to cite this URL: Gunti H, Maruthiramaih V. V, Shivanandappa T. B. Bio-degradation of Azo Dye Acid Orange-10 by a New Isolate of Bacillus subtilis Isolated from Soil Sample around Textile Industry in South Karnataka.Biosci Biotech Res Asia 2020;17(4). Available from: https://bit.ly/3ivRTt2 |
Introduction
Synthetic dyes are important industrial coloring agents. Dyes are characterized by chromophore bunches in their compound structures and are primarily grouped as azo colors, anthraquinone colors, phthalocyanine dyes etc.1 They are used as scatter colors for polyester and reactive dyes for cotton. The absorbance spectrum of azo dyes lies in the visible region,which can be attributed to theircommon structure, consisting of at least one azo bond (- N=N-).2
Globally, 2.8×105 tons of colored chemical compounds are allowed to drain into the ecological water system each year.3 Azo compounds are the most widely useddyes, on account of their simpler synthesis, concoction dependability and the decent variety of hues accessible when contrasted with normal dyes.4 They are generally utilized intextiles, leather,pharmaceutical,paper, food, makeup, and pharmaceutical industries. Analyses indicate that 10-15% of the coloring agents utilized in coloring process do not bind with the textile strands and get released into effluents.5
Discharge of the dye-containing wastes from different industrial practices into surrounding water bodies is of significant concern, and has many unfavorable consequences, including diminished aquatic photosynthesis, exhaustion of available DO and harmful impact on different life forms. These colored effluents not only cause a disagreeable appearance of water bodies, but alsorelease poisonous colorless amines by breakdown of the dyes, which aremutagenic, carcinogenic and capable of causing other health hazards.6, 7
Different research activities uncovered the utilization of physico-chemical methodology strategies for removal of dyes from the effluents.8,9 These techniques incorporate use of adsorption agents, particle pair extractions, coagulation, and chemical processes, which also posesignificant connected issues, for example, high expense and production of a lot of slime after treatment, disposal of which is cumbersome and hazardous.
Various studies available on utilization of microorganisms for dye degradation in effluents, recommend it to be an eco-friendly and economically viable technique.10, 11 The advantages of microbial/enzymatic methodology also include less slime production and hence more practical.12,13 With this backdrop, the present study was aimed at studying the effect of various physical parameters on azo dye Acid orange-10 decolorization by a new isolate of Bacillus subtilis.
Materials and Methods
Chemicals required
The Acid orange-10 dye was obtained from Shailaja textile industry, Sholapur Maharashtra, India. Dehydrated culture medium obtained from Hi-media and other chemicalsand reagents used are of analytical grade.
Acid Orange-10 Decolorizing Bacteria-Isolation, Screening and Identification
The dye decolorizing bacteria were isolated from the soil samples in and around textile industrial area in South Karnataka. 10 gm of soil sample was suspended in 100 ml of complete medium broth supplemented with Acid orange-10 (100mg/L) individually and acclimatized for 5 days at 30°C at 150 rpm. Bushnell and Haas medium (BHM) containingMgSO4-0.2, K2HPO4-1.0, CaCl2– 0.02, FeCl3-0.05, NH4NO3-1.0 (g l-1) supplemented with glucose and yeast extract (0.1% and 0.05% w/v) respectively,at pH 7.0.
The dye decolorizing bacteria were isolated on BHM agar (pH 7.0) containing Acid orange-10 (100mg/L.) from acclimatized soil sample using serial dilution. All the bacterial isolates were studied by inoculating them in complete medium broth supplemented with dye. The inoculated liquid broth medium was incubated at 30°C /37°C under shaking condition at 150 rpm for 1-5 days. The decolorization was visually observed. The isolates showing considerable decolorization of the dyes were selected for further investigation. Morphological and biochemical tests for identification of the selected bacterial isolates were based on the Bergey’s Manual of Systematic Bacteriology.14
Decolorization Assay
The decolorized supernatant (aliquots of 2 ml each) at regular intervals of time were collected and subjected to centrifugation at 10000 rpm for 10 min to remove any cells to avoid interference with the spectroscopic measurement. The supernatant was used for spectrophotometric analysis (Shimadzu 1900) at 300-700 nm. The absorbance maximum of Acid orange10 is at 480nm (Fig.1). The efficiency of decolorization was determined using the given formula
D= [A0-A1)/A0 A1] x 100
Where,
D – Decolorization (%)
A0 – Initialabsorbance before decolorization
A1 – Final absorbance after decolorization
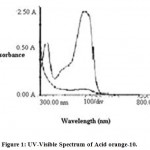 |
Figure 1: UV-Visible Spectrum of Acid orange-10. |
Identification of Metabolites
The culture medium after complete decolorization of Acid orange-10 was subjected to centrifugation at 10,000 rpm for 15 min. The 200 ml of the supernatant was taken after bringing pH to 7 and extracted twice with diethylether (500 ml). The extraction was repeated after adjusting the pH of the remaining aqueous layer was brought to 2, using 1N HCl. The acidic and alkaline extracts were combined and evaporated using anhydrous Na2SO4 under a reduced pressure at 30oC. Then the residue was suspended in 0.5ml of methanol and then subjected to TLC.
Thin Layer Chromatography (TLC)
TLC of extracted compounds was performed to identify the components of the decolorized medium. An aqueous slurry of silica gel- G (40% w/v) with binder was coated over a glass plate (200×100×2 mm) dimension for TLC. 10 µl each of standard Acid orange-10 dye and of the extracted fractions was loaded on the glass plate coated with silica gel.The solvent system used for developing the TLC plateconsisted of propanol: water: acetic acid (80:19:01). The chromatogram was developed by exposing the plate to Iodine vapors. The diazotization and carbylamines test were used for the identification of metabolites.
Standardization of Various Operational and Environment Conditions
Different operational and environmental conditions were standardized, by varying only oneparameter, maintaining the other constant at a time. Following parameters were standardized and their effects were observed on decolorization of Acid orange10. Effect of temperature on decolorization was studied between 20-500C, different pH levels ranging from 4-10 at an interval of 0.5, different inoculum sizes ranging from 1-10% in 100 ml of culture media containing 30 ppm dye and different dye concentration 50-1000 mg/L and shaking rpm in the range of 50-200 rpm.
Bushnell and Haas medium as mentioned earlier was used for all the studies
Results
Isolation, Screening and Identification of Acid Orange-10 Decolorizing Bacteria
Bacterial species capable of decolorizing Acid Orange-10 were isolated from different sources using Bushnell-Hass medium containing 100 ppm dye at 37oC. The isolates efficient in decolorizing Acid orange-10 were selected through visual observation up to 72hrs of incubation. The isolate which took shortest period of 16 hrs for complete decolorization was used for further studies. A series of tests was performed according to the Bergey’s manual to identify the microorganism, the results of which are presented in Table 1. According to the results obtained in these tests, the isolate was identified as Bacillis subtilis.
Table 1: Morphological and Biochemical Characteristics shown by the new isolate of Bacillus subtilis.
| Test | Result |
| Gram’s Staining | + |
| Shape | Rod |
| Motility | Motile |
| Indole Production | – |
| Methyl Red | – |
| Voges-Prausker | + |
| Citrate utilization | + |
| Catalase | + |
| Oxidase | – |
Identification of Metabolic Intermediates:
The products of dye degradation were separated on TLC plates using solvent system propanol: water: acetic acid (80:19:01). The separated products of TLC plates when exposed to iodine vapors showed two degraded products with 0.75 and 0.56 Rf values, whereas control Rf value is 0.50 (Figure 2). Each degraded product was analyzed by diazotization and carbolamine test, both the degraded compounds with Rf values of 0.75, 0.56 gave positive results, identified and confirmed as aromatic amines.
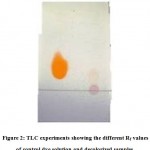 |
Figure 2: TLC experiments showing the different Rf valuesof control dye solution and decolorized samples. |
Optimization of Different Parameters for Acid orange-10 Decolorization
Effect of Incubation Time
The samples were removed at different intervals of time during incubation viz. 6, 12, 18 and 24 hrs and scanned for λmax of Acid orange-10. The results indicated a gradual disappearance of λmax peak(at 480nm) to complete disappearance in 24 hrs. These observations indicate the disappearance of the Acid orange-10dye in its original form and could be due to modification or breakdown of dye by the bacterium.
Effect of shaking
Aeration may be either favorable or inhibitive towards microbial decolorization of dyes. To study the effect of aeration on decolorization of Acid orange10, Bacillus subtilis cells were cultured in nutrient broth containing dye under static and shaking cultural conditions; while the time taken for complete decolorization at static conditions was 24 hrs, at a shaking speed of 50 rpm, it took 36 hrs for complete decolorization; further increase in speed of shaking to 100, 150 and 200 rpm, the time taken for 100% decolorization was increased to 48 hrs, 72 hrs and 96 hrs respectively, indicating that the decolorization process was inhibited upon increase of aeration (Figure 3).
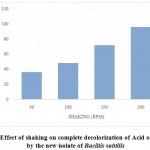 |
Figure3: Effect of shaking on complete decolorization of Acid orange-10 by the new isolate of Bacillis subtilis |
Effect of Temperature
The temperature between 35-450C was found to be an optimum range for the complete decolorization of Acid orange10. At 400C the complete decolorization (i.e.100%) was found with in the shortest time of 22hrs, hence 40oC could be taken as the optimum. At 200C the decolorization of Acid Orange10 was very slow and it took almost 72 hrs for 100%decolorization. At 600C the time required increased to 84hrs while above 600C complete decolorization did not occur (Figure 4).
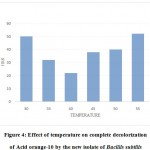 |
Figure 4: Effect of temperature on complete decolorization of Acid orange-10 by the new isolate of Bacillis subtilis |
Effect of pH
The results of the present study depict that this isolate was capable of 100% decolorization of Acid orange10 at a wide range of pH i.e .6.00 to 9.50. The optimum decolorization occurred at pH 8.50 wherein 300 ppm of the dye was decolorized within 24 hours. At pH 5.5 and below and also at pH 10, 100% decolorization could not be achieved (Fig.5).
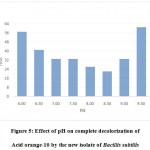 |
Figure 5: Effect of pH on complete decolorization of Acid orange-10 by the new isolate of Bacillis subtilis. |
Effect of Dye Concentration
As the concentration of Acid orange-10 increases the time taken for the complete decolorization also increases. The bacterium decolorized 100 ppm of dye completely (i.e.100%) in 12hrs of incubation whereas 100% decolorization of 200 ppm of Acid orange-10 dye was found at 24 hrs. Further increase in dye concentration to 300, 400, 500, 600 and 700 ppm, the time required for complete decolorization of the azo dye increased to 32, 48, 56, 62 and 72hrs respectively. The isolate decolorized the Acid orange-10 completely up to a maximum concentration of 700 ppm, but the time taken was 72 hrs. A dye concentration above 700 ppm was not completely decolorized even after extended incubation period (Fig.6).
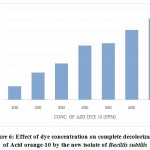 |
Figure 6: Effect of dye concentration on complete decolorization of Acid orange-10 by the new isolate of Bacillis subtilis |
Effect of Inoculum size
The study of effect of the inoculum size of the newly isolated Bacillus subtilis on the decolorization of Acid Orange-10 (300 ppm) indicated that increase in the inoculum size from 1-10% progressively, decreased the time required for the complete decolorization. The minimum inoculum size of 1% required 37hrs for 100% decolorization of Acid orange-10, and as the inoculum size increased the time taken for complete decolorization of Acid orange-10 dye was decreased from 37hrs at 1% to 17hrs at 10% of inoculum size (Fig.7).
 |
Figure 7: Effect of inoculum size on complete decolorization of Acid orange-10 by the new isolate of Bacillis subtilis |
The isolate showed the characteristics of Bacillis subtillis in the assays performed for identification of the organism as shown in the Table 1. Similar kind of observations were reported by Ponraj et al.15
Decolorization and degradation of azo dyes can be either due to biosorption (adsorption on the microbial biomass) or enzymatic biodegradation. Decolorization of Acid orange10 by Bacillus subtilis in our study was not due to adsorption, because when bacterial cell mass was collected and treated with solvents methanol or chloroform, there was no release of color from the cells. Appearance of aromatic amines in the TLC of decolorized culture supernatant indicates degradation of the dye by the bacterium. Biodegradation of textile azo dyes has been suggested as the best model because of removal of practically all of the dye with minimal disadvantages presented by the physico-chemical procedures. Various studies also emphasize on the utilization of microorganisms for dye containing wastewater treatment as an eco-friendly and cost-effective technique.10, 11
Study of available literature study shows that bacterial degradation of azo dyes involves azo-reductase mediated disruption of azo bond (- N=N-) under anaerobic conditions, leading to formation of colorless, hazardous aromatic amines, which could be removed further.16, 17 Formation of colorless aromatic amines by reductive cleavage of -N=N- (azo) bond is suggested to be the initial step in bacterial degradation of dyes.
Earlier studies have reported Bacillus subtilis18, Aeromonas hydrophilia 19 and Bacillus cereus20 culture capable of degrading azo dyes. Lalnunhlimi and Krishnaswamy report a consortium of different Bacillus species fit for decolorization of direct blue 151 and 31 up to 95%.21 Presence of aromatic amines in TLC in the current study is an indication that decolorization was due to degradation of the Acid orange-10 dye into intermediate products.
Time taken by the bacterial isolatein the present study was much less than previous reports. Jothimani and Prabhakaran report that Pseudomonas and Bacillus species can remove 59% dye from industrial effluents in a period of 14 days.22 Bacterial decolorization of direct blue dye by Bacillus sp. ETL-1979, with an incubation period of 168 hr was reported by Shah et al. 23 96.4% decolorization of black WNN (100mg/L) was obtained in 48 hr by Paenibacillus alvei MTCC 10625.24 Decolorization of direct blue 151 at a concentration of 200 mg/L was reported to be 95.25% in 5 days.21
Inhibition of dye decolorization at higher speed of shaking indicates that decolorization process was inhibited in presence of higher concentration of oxygen whereas lower rpm ort static condition i.e. lower concentration of oxygen was required to enhance decolorization.The reports of current study are in concurrence with results reported by Verma and Madamwar wherein the authors have reported that under static conditions decolorization was high and under agitation, decolorization was negligable.25
Azoreductase is a cytoplasmic enzyme with low specificity, and is responsible for reduction of azo bonds in the azo dyes, degrading them to their corresponding amines. The reaction is slowed down or is inhibited by the presence of O2, since it is a strong terminal electron acceptor compared to the azo groups during the oxidation of reduced electron carriers like NADH.26,27 Under static conditions, electrons required for azo bond cleavage are readily accessible to the enzyme from NADH; while under shaking, the availability of O2 prevents the azoreductase from receiving the electrons.28,29
Kumar and Sawhney reported that Bacillus subtilis (RA 29) caused decolorization of Congo red to the extent of 95.67% at 37°C.30 Bayoumi et al., had reported maximum bacterial decolourization of Acid orange 7 and Direct blue 75occurs at an optimum incubation temperature of 35°C.31
The findings of our study with respect to the effect of pH on decolorization of acid orange-10 are in agreement with other studies in which decolorization of Methyl Red was maximal in pH range of 6-8 by a strain of Micrococcus32. Removal of Acid red 2 and Acid orange 7 by Bacillus 33 and of Acid Orange by a strain of Staphylococcus hominis was also found to be maximum in the range of 6-8 pH.34 Bayoumi et al., reported that neutral to moderately alkaline pH enhance the removal of azo dyes.31
Karunya et al., reported that the dye concentration at 200mg/l was completely decolorized in 48 hrs by Pseudomonas aeruginosa, but beyond this, the decolorization was not effective.35
The percentage decolorization of azo dye has linear relationship with inoculum size. The results of the current study are in corroboration with the reports of Kumar et al.36 Gurulakshmi et al., reported that maximal decolorization by B.subtilis strain was achieved at 20% inoculum size.37
Conclusion
Under static condition the new isolate of Bacillus subtilis from soil sample contaminated with textile effluent could completely decolorize the Acid orange-10 mono azo dye. Optimization of various parameters like temperature, pH, inoculum size, aeration and incubation period were done to enhance the decolorization process of Acid orange-10. The currentlyisolated Bacillus subtilis could efficiently decolorize the dye even at higher concentrations up to 700 ppm. Hence the new isolate of the microorganism Bacillus subtilis is found to be potential candidate for Acid orange-10 azo dye decolorization in textile effluents. Further studies can help to successfully employ this bacterium for treating the textile effluents before they are released into the surroundings.
Acknowledgement
The authors’acknowledge the support of all people who have helped to carry out this research work. We also thank Maharani’s Science College for Women, palace Road, Bangalore, for providing infrastructure.
Funding source
This is a self-funded research work.
Conflict of Interest
The authors declare no conflict of interest.
References
- Singh P, Iyengar L and Pandey A. Bacterial decolorization and degradation of azo dyes. In: Singh, S.N., (ed.) Microbial Degradation of Xenobiotics. Springer, Heidelberg Dordrecht London New York, 2012; pp.101-131.
CrossRef - Chang JS, Chou C, Lin YC, Lin PJ, Ho JY, Hu TL. Kinetic characteristics of bacterial azo-dye decolorization by Pseudomonas luteola. Water Research. 2001;35(12):2841-50.
CrossRef - Jin XC, Liu GQ, Xu ZH, Tao WY. Decolorization of a dye industry effluent by Aspergillus fumigatus Applied Microbiology and Biotechnology. 2007;74(1):239-43.
CrossRef - Chang JS, Chen BY, Lin YS. Stimulation of bacterial decolorization of an azo dye by extracellular metabolites from Escherichia coli strain NO3. Bioresource Technology. 2004;91(3):243-8.
CrossRef - Asad S, Amoozegar MA, Pourbabaee A, Sarbolouki MN, Dastgheib SM. Decolorization of textile azo dyes by newly isolated halophilic and halotolerant bacteria. Bioresource Technology. 2007;98(11):2082-8.
CrossRef - Mugdha A, Usha M. Enzymatic treatment of wastewater containing dyestuffs using different delivery systems. Sci Rev Chem Commun. 2012;2(1):31-40.
- Xu M, Guo J, Cen Y, Zhong X, Cao W, Sun G. Shewanelladecolorationis sp. nov., a dye-decolorizing bacterium isolated from activated sludge of a waste-water treatment plant. J. Systematic and Evolutionary Microbiology. 2005;55(1):363-8.
CrossRef - Golob V, Vinder A, Simonič M. Efficiency of the coagulation/flocculation method for the treatment of dyebath effluents. Dyes and Pigments. 2005;67(2):93-7.
CrossRef - López-Grimau V, Gutierrez MC. Decolorization of simulated reactive dyebath effluents by electrochemical oxidation assisted by UV light. Chemosphere. 2006;62(1):106-12.
CrossRef - Vitor V, Corso CR. Decolorization of textile dye by Candida albicans isolated from industrial effluents. Industrial Microbiology &Biotechnology. 2008;35(11):1353-7.
CrossRef - Pajot HF, Delgado OD, de Figueroa LI, Farina JI. Unraveling the decolorizing ability of yeast isolates from dye-polluted and virgin environments: an ecological and taxonomical overview. Antonie van Leeuwenhoek. 2011;99(3):443-56.
CrossRef - Singh S, Chatterji S, Nandini PT, Prasad AS, Rao KV. Biodegradation of azo dye Direct Orange 16 by Micrococcusluteus strain Inter. J of Environmental Science and Technology. 2015; 12(7):2161-8.
CrossRef - Sari IP and Simarani K. Decorization of selected azo dye by Lysinibacillus fusiformis W1B6: Biodegradation optimization, isotherm, and kinetic study biosorption mechanism. Adsorption Science and Technology.2019; 37(5-6):492-508.
CrossRef - Sneha J, Gomashe AV, Sonal A. Decolorization potential of Bacillus sp. for removal of synthetic textile dyes. J of Current Microbiology and Applied Sciences. 2014; 3(12):83-8.
- Ponraj M, Gokila K, Zambare V. Bacterial decolorization of textile dye-Orange 3R. J Advanced Biotechnology and Research. 2011; 2(1):168-77.
- Van der Zee FP, Villaverde S. Combined anaerobic–aerobic treatment of azo dyes—a short review of bioreactor studies. Water Research. 2005; 39(8):1425-40.
CrossRef - Joshi T, Iyengar L, Singh K, Garg S. Isolation, identification and application of novel bacterial consortium TJ-1 for the decolorization of structurally different azo dyes. Bioresource Technology. 2008; 99(15):7115-21.
CrossRef - Horitsu H, Takada M, Idaka E, Tomoyeda M, Ogawa T. Degradation of p-Aminoazobenzene by Bacillus subtilis. Eur J Applied Microbiology and Biotechnology. 1977; 4(3):217-24.
CrossRef - Idaka E, Ogawa T, Horitsu H, Tomoyeda M. Degradation of azo compounds by Aeromonas hydrophila var. 24B. J the Society of Dyers and Colorists. 1978; 94(3):91-4.
CrossRef - Wuhrmann K, Mechsner KL, Kappeler TH. Investigation on rate—Determining factors in the microbial reduction of azo dyes. Eur J Applied Microbiology and Biotechnology. 1980; 9(4):325-38.
CrossRef - Lalnunhlimi S, Krishnaswamy V. Decolorization of azo dyes (Direct Blue 151 and Direct Red 31) by moderately alkaliphilic bacterial consortium. Brazilian JMicrobiology. 2016; 47(1):39-46.
CrossRef - Jothimani P, Prabakaran J. Influence of bacterial system on the decolorization of dye effluent under enrichment techniques. In State Level Seminar in Recent Developments in Applied Microbiology, Tamil Nadu Agricultural University. Coimbatore, pp-25-26 1998.
- Sha MP, Patel KA, Nair SS, Darji AM. An application of response surface methodology in microbial degradation of azo dye by Bacillus subtillis ETL-1979. Am J Microbiol Res. 2014; 2:24-34.
CrossRef - Pokharia A, Ahluwalia SS. Decolorization of Xenobiotic Azo Dye-Black WNN by Immobilized Paenibacillus alvei MTCC 10625. Int J Environ BioremedBiodegrad. 2016; 4:35-46.
- Verma P, Madamwar D. Decolourization of synthetic dyes by a newly isolated strain of Serratia marcescens. World J Microbiology and Biotechnology. 2003; 19(6):615-8.
CrossRef - Chang JS, Lin YC. Fed-batch bioreactor strategies for microbial decolorization of azo dye using a Pseudomonas luteola strain. Biotechnol Progress. 2000; 16:979–85.
CrossRef - Misal SA, Gawai KR. Azoreductase: a key player of xenobiotic metabolism. Bioprocess. 2018. 5:17.
CrossRef - Stolz A. Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol. 2001; 56:69–80.
CrossRef - Mahmood S, Khalid A, Arshad MG, Mahmood T and Crowley DE. Detoxification of azo dyes by bacterial oxidoreductase enzymes. Critical Reviews in Biotechnology. 2015; 36(4): 1-13.
CrossRef - Kumar A, Sawhney R. Identification of Bacillus subtilis subsp. subtilis “RA-29”, a congo red decolorizer using 16S rDNA sequencing. Researcher. 2011; 3:18-22.
- Bayoumi RA, Musa SM, Bahobil AS, Louboudy SS, El-Sakawey TA. Bio decolorization and biodegradation of azo dyes by some bacterial isolates. J Appl Environ Biol Sci. 2010; 1(1):1-25.
- Olukanni DO, Osuntoki AA, Gbenle GO. Decolourization of azo dyes by a strain of Micrococcus isolated from a refuse dump soil. Biotechnology. 2009; 8(4):442-8.
CrossRef - Sneha SJ, Gomashe AV. Bioremediation of textile azo dyes by newly isolated Bacillus sp. from dye contaminated soil. International Journal of Biotechnology and Biochemistry. 2017; 13(2):147-153.
- Singh RP, Singh PK, Singh RL. Bacterial decolorization of textile azo dye acid orange by Staphylococcus hominis Toxicology Int. 2014; 21(2):160.
CrossRef - Karunya A, Rose C. Studies on degradation of textile azo dye, Mordant Black 17 using Pseudomonas aeruginosa SUB 7, isolated from textile effluent. Int J Applied Bioengineering. 2013; 7(1).
CrossRef - Kumar K, Dastidar MG, Sreekrishnan TR. Effect of process parameters on aerobic decolorization of reactive azo dye using mixed culture. World Acad SciEng Technol. 2009; 58:952-5.
- Gurulakshmi M, Sudarmani DN, Venba R. Biodegradation of leather acid dye by Bacillus subtilis.Adv Biotech. 2008; 7:12-9.

This work is licensed under a Creative Commons Attribution 4.0 International License.





